作物杂志,2020, 第6期: 23–29 doi: 10.16035/j.issn.1001-7283.2020.06.004
玉米秸秆消化率全基因组关联分析
林淼1( ), 张秋芝1, 史利玉1, 刘蓓2, 王红武2, 潘金豹1(
), 张秋芝1, 史利玉1, 刘蓓2, 王红武2, 潘金豹1( )
)
- 1北京农学院,102206,北京
2中玉金标记(北京)生物技术股份有限公司,102206,北京
Genome-Wide Association Study of Digestibility of Straw in Maize
Lin Miao1( ), Zhang Qiuzhi1, Shi Liyu1, Liu Bei2, Wang Hongwu2, Pan Jinbao1(
), Zhang Qiuzhi1, Shi Liyu1, Liu Bei2, Wang Hongwu2, Pan Jinbao1( )
)
- 1Beijing University of Agriculture, Beijing 102206, China
2Zhongyu Golden Marker (Beijing) Biotechnology Incorporated Company, Beijing 102206, China
摘要:
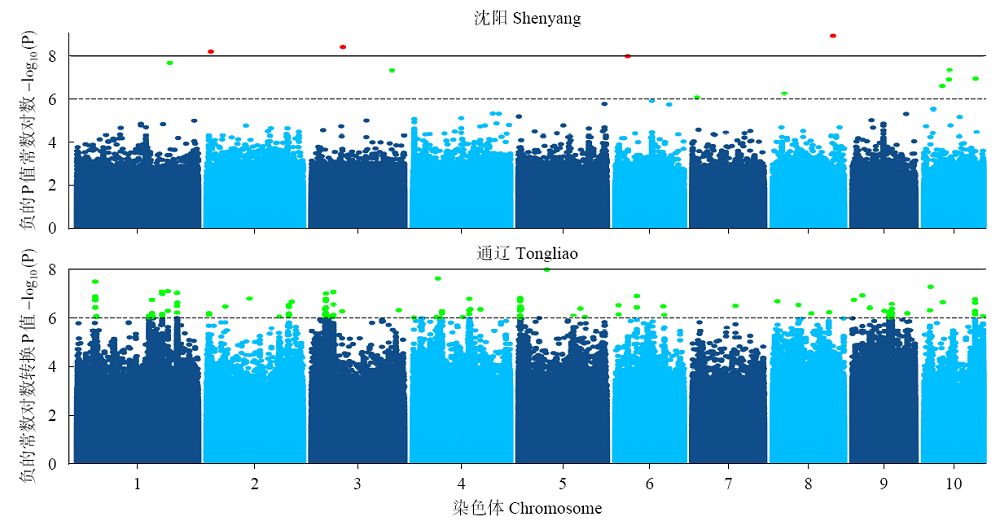
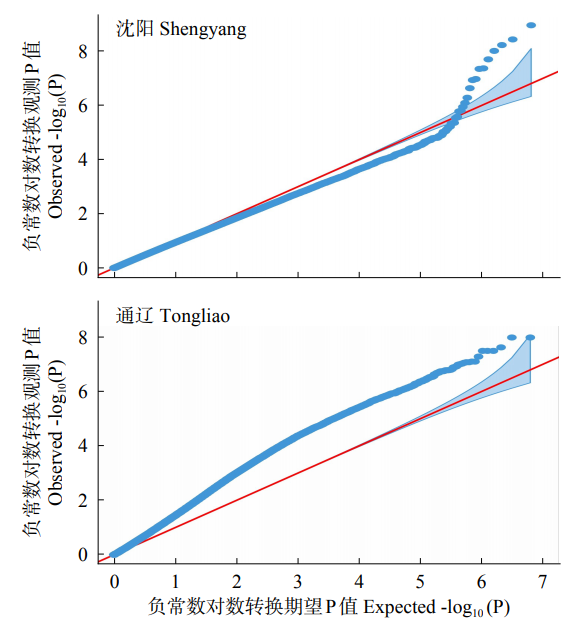
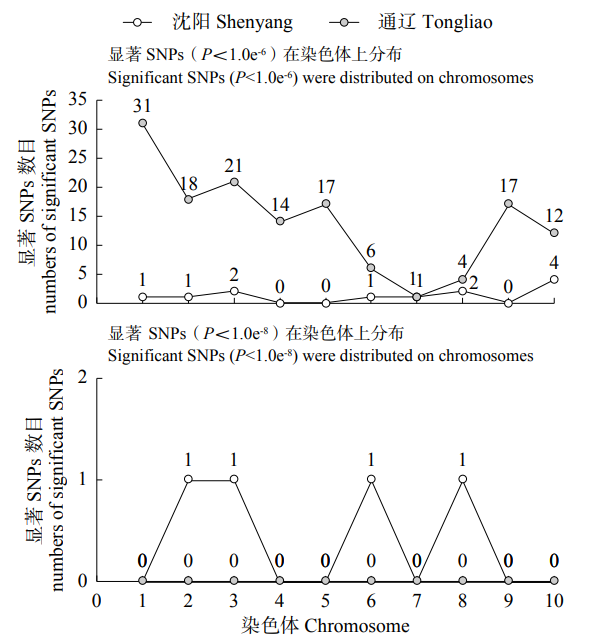
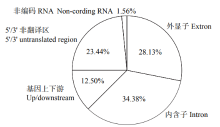
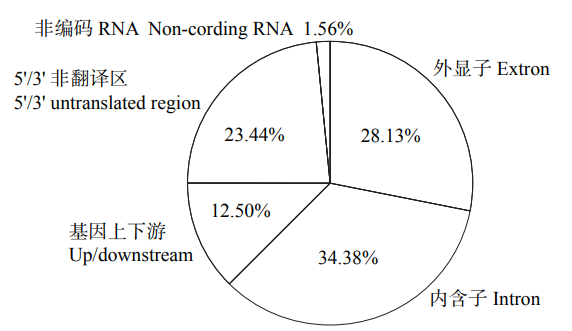
48h体外干物质消化率(48h in vitro dry matter digestibility, 48h IVDMD)是衡量青贮玉米品质的重要指标。为了初步探究玉米秸秆消化率的分子遗传机理,以341份玉米自交系为材料,于2018年在沈阳和通辽种植,收获后测定秸秆48h IVDMD。利用全基因组重测序获得的6 276 612个高质量SNPs进行全基因组关联分析,共检测到153个与玉米秸秆消化率显著相关的SNPs位点(P<1.0×10-6),4个SNPs显著水平在P<1.0×10-8以上;共找到38个秸秆消化率的候选基因,主要涉及细胞生长发育、防御反应和信号转导等生物学功能。
| [1] | 潘金豹, 张秋芝, 郝玉兰, 等. 我国青贮玉米育种的策略与目标. 玉米科学, 2002,10(4):3-4. |
| [2] | Yves B, Carine G, Deborah G, et al. Genetic variation and breeding strategies for improved cell wall digestibility in annual forage crops. A review. Animal Research, 2003,52:193-228. |
| [3] | 白琪林, 陈绍江, 董晓玲, 等. 近红外漫反射光谱法测定玉米秸秆体外干物质消化率. 光谱学与光谱分析, 2006,26(2):271-274. |
| [4] | 张吉鹍, 邹庆华, 张䴔, 等. 奶牛粗饲料纤维品质的综合评定研究. 中国奶牛, 2009(1):20-22. |
| [5] |
Li K, Wang H W, Hu X J, et al. Genetic and quantitative trait locus analysis of cell wall components and forage digestibility in the Zheng58 × HD568 maize RIL population at anthesis stage. Frontiers in Plant Science, 2017,8:1472.
doi: 10.3389/fpls.2017.01472 pmid: 28883827 |
| [6] | 王琪. 利用连锁和关联分剖析玉米茎秆细胞壁组分及消化性状的遗传基础. 北京:中国农业科学院, 2015. |
| [7] | Wang H W, Han J, Sun W T, et al. Genetic analysis and QTL mapping of stalk digestibility and kernel composition in a high-oil maize mutant (Zea mays L.). Plant Breeding, 2010,129:318-326. |
| [8] | Méchin V, Argillier O, Hebert Y, et al. Genetic analysis and QTL mapping of cell wall digestibility and lignification in silage maize. Crop Science, 2001,41:690-670. |
| [9] |
Wang H W, Li K, Hu X H, et al. Genome-wide association analysis of forage quality in maize mature stalk. BMC Plant Biology, 2016,16:227.
pmid: 27769176 |
| [10] | 杨小红, 严建兵, 郑艳萍, 等. 植物数量性状关联分析研究进展. 作物学报, 2007,33(4):523-530. |
| [11] | Huang X Y, Han B. Natural variations and genome-wide association studies in crop plants. annual. review. Plant Biology. 2014,65:531-551. |
| [12] | 贺城, 杨增玲, 黄光群, 等. 可见/近红外光谱分析秸秆-煤混燃物的秸秆含量. 农业工程学报, 2013,29(17):188-195. |
| [13] | 陈丽丽, 张鹏, 黄新, 等. 玉米基因组DNA提取及浓度测定方法评价. 生物技术通报, 2011(12):70-74. |
| [14] |
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 2009,25(14):1754-1760.
doi: 10.1093/bioinformatics/btp324 pmid: 19451168 |
| [15] |
Raj A, Stephens M, Pritchard J K. FastSTRUCTURE:variational inference of population structure in large SNP data sets. Genetics, 2014,197(2):573-589.
doi: 10.1534/genetics.114.164350 |
| [16] | 刘晓磊. 一种交替运用固定效应和随机效应模型优化全基因组关联分析的算法开发. 武汉:华中农业大学, 2015. |
| [17] |
Wu X, Li K Y, Zheng Y X, et al. Genetic discovery for oil production and quality in sesame. Nature Communications, 2015,6:8609.
doi: 10.1038/ncomms9609 pmid: 26477832 |
| [18] |
Barrett J C, Fry B, Maller J, et al. Haploview:Analysis and visualization of LD and haplotype maps. Bioinformatics, 2005,21(2):263-265.
doi: 10.1093/bioinformatics/bth457 pmid: 15297300 |
| [19] |
Wang K, Li M H, Hakonarson. ANNOVAR:functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research, 2010,38(16):e164.
doi: 10.1093/nar/gkq603 pmid: 20601685 |
| [20] |
Cristina B, Isabel P, Luísa R. Gene expression regulation by upstream open reading frames and human disease. PLoS Genetics, 2013,9(8):e1003529.
doi: 10.1371/journal.pgen.1003529 pmid: 23950723 |
| [21] |
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit:a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 2010,20(9):1297-1303.
pmid: 20644199 |
| [22] | 曹运琳, 邢梦云, 徐昌杰, 等. 植物黄酮醇生物合成及其调控研究进展. 园艺学报, 2018,45(1):177-192. |
| [23] | 张瑜. 拟南芥FRIGIDA-LIKE蛋白家族成员AtFRL4基因及其上游调控序列的克隆与转化研究. 南京:南京农业大学, 2012. |
| [24] | Roe J L, Durfee T, Zupan J R, et al. TOUSLED is a nuclear serine/threonine protein kinase that requires a coiled-coil region for oligomerization and catalytic activity. Journal of Biological Chemistry, 1997,272(9):5838-5845. |
| [25] | Mark H L, Lambermon Y F, Lorković Z J. UBA1 and UBA2,two proteins that interact with UBP1,a multifunctional effector of pre-mRNA maturation in plants. Molecular and cell Biology, 2002,22(12):4346-4357. |
| [26] |
Pagant S, Bichet A, Sugimoto K, et al. KOBITO1 encodes a novel plasma membrane protein necessary for normal synthesis of cellulose during cell expansion in Arabidopsis. The Plant Cell, 2002,14(9):2001-2013.
doi: 10.1105/tpc.002873 pmid: 12215501 |
| [27] | Xin W, Yan J J, Bao C, et al. Glycosyltransferase-like protein ABI8/ELD1/KOB1 promotes Arabidopsis hypocotyl elongation through regulating cellulose biosynthesis. Plant Cell & Environment, 2014,38:411-422. |
| [28] | 解敏敏, 晁江涛, 孔英珍. 参与木葡聚糖合成的糖基转移酶基因研究进展. 植物学报, 2015,50(5):644-651. |
| [29] |
Chou Y H, Gennady P, Olga A Z. Xyloglucan Xylosyltransferases XXT1,XXT2,and XXT5 and the Glucan Synthase CSLC4 form golgi-localized multiprotein complexes. Plant Physiology, 2012,159(4):1355-1366.
doi: 10.1104/pp.112.199356 pmid: 22665445 |
| [30] |
Wang C, Li S, Sophia N, et al. Mutation in xyloglucan 6-xylosytransferase results in abnormal root hair development in Oryza sativa. Journal of Experimental Botany, 2014,65(15):4149-4157.
doi: 10.1093/jxb/eru189 pmid: 24834920 |
| [31] |
Vorwerk S, Somerville S, Somerville C. The role of plant cell wall polysaccharide composition in disease resistance. Trends in Plant Science, 2004,9(4):203-209.
doi: 10.1016/j.tplants.2004.02.005 |
| [32] |
T. Heitz, D R. Bergey, C A. Ryan. A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding,systemin,and methyl jasmonate. Plant Physiology, 1997,114(3):1085-1093.
doi: 10.1104/pp.114.3.1085 pmid: 9232884 |
| [33] | 宗娜, 阎云花, 王琛柱. 昆虫对植物蛋白酶抑制素的诱导及适应机制. 昆虫学报, 2003(4):533-539. |
| [34] |
Chen J, Burke J J, Velten J, et al. FtsH11 protease plays a critical role in Arabidopsis thermotolerance. The Plant Journal, 2006,48(1):73-84.
doi: 10.1111/j.1365-313X.2006.02855.x pmid: 16972866 |
| [35] | 陈发晶, 谭蕊, 黄萌雨, 等. 类枯草杆菌蛋白酶在植物和病原物互作中的研究进展. 分子植物育种, 2018,16(10):3146-3153. |
| [36] | Albertsen M, Fox T, Leonard A, et al. Cloning and use of the ms9 gene from maize:US20160024520A1. 2015-12-23. |
| [37] | 戴毅, 高莹莹, 黄泽峰, 等. 玉米R2R3型转录因子家族生物学功能综述. 江苏农业科学, 2013,41(3):6-8. |
| [1] | 董怀玉, 董智, 刘可杰, 王丽娟, 刘培斌, 侯志研. 不同秸秆还田模式对玉米主要病害发生为害的影响[J]. 作物杂志, 2020, (6): 104–108 |
| [2] | 菅立群, 张翼飞, 杨克军, 王玉凤, 陈天宇, 张继卫, 张津松, 李庆, 刘天昊, 肖珊珊, 彭程, 王宝生. 播种至苗期不同阶段低温对玉米幼苗生长及生理抗性的影响[J]. 作物杂志, 2020, (6): 61–68 |
| [3] | 任云, 刘静, 李哲馨, 李强. 玉米幼苗响应低铁胁迫的根系形态与干物质积累特征[J]. 作物杂志, 2020, (6): 69–79 |
| [4] | 黄少辉, 杨军芳, 刘学彤, 杨云马, 邢素丽, 韩宝文, 刘孟朝, 贾良良, 何萍. 长期小麦秸秆还田对壤质潮土磷素含量及磷盈亏的影响[J]. 作物杂志, 2020, (6): 89–96 |
| [5] | 张刚, 张世洁, 王德建, 俞元春. 稻麦两熟制下秸秆还田模式的产量和经济效益分析[J]. 作物杂志, 2020, (6): 97–103 |
| [6] | 阎晓光, 杜艳伟, 李洪, 董红芬, 李爱军, 王国梁, 周楠. 玉米籽粒水分的快测法误差分析[J]. 作物杂志, 2020, (5): 170–173 |
| [7] | 吴琼, 丁凯鑫, 余明龙, 黄文婷, 左官强, 冯乃杰, 郑殿峰. 新型植物生长调节剂B2对玉米光合荧光特性及产量的影响[J]. 作物杂志, 2020, (5): 174–181 |
| [8] | 吴伟华, 柳家友, 袁刘正, 闫海霞, 付家锋, 王会强, 王蕊, 李腾, 刘康. 孕穗期高温逆境对玉米杂交种的影响分析[J]. 作物杂志, 2020, (5): 59–64 |
| [9] | 曾艳华, 谢和霞, 江禹奉, 周锦国, 谢小东, 周海宇, 谭贤杰, 覃兰秋, 程伟东. 基于SNP标记的爆裂玉米农家品种遗传多样性[J]. 作物杂志, 2020, (5): 65–70 |
| [10] | 李忠南, 王越人, 张艳辉, 邬生辉, 曲海涛, 许正学, 李光发. 玉米DH系15D969超多穗行数的遗传分析[J]. 作物杂志, 2020, (5): 88–92 |
| [11] | 张丛卓, 邢丽君, 李南, 闫枫, 田怀东. 玉米胚乳中最大量蛋白体的提纯与体外消化[J]. 作物杂志, 2020, (5): 98–102 |
| [12] | 王莉, 王作平, 张中保, 白玲, 吴忠义. 玉米早期籽粒中强表达启动子的筛选[J]. 作物杂志, 2020, (4): 114–120 |
| [13] | 范园园, 吴海梅, 逄蕾, 路建龙, 夏博文, 杨旭海. 基于Meta分析评价秸秆覆盖对我国北方半干旱区不同生态区域小麦产量的影响[J]. 作物杂志, 2020, (4): 143–149 |
| [14] | 李强, 孔凡磊, 袁继超. 年际气象差异对西南丘陵区玉米物质积累与产量的影响[J]. 作物杂志, 2020, (4): 150–157 |
| [15] | 郑飞, 汪丽霞, 刘瑞响, 孔令杰, 陈艳萍, 袁建华, 崔亚坤. 淹水胁迫下玉米自交系苗期的形态生理差异[J]. 作物杂志, 2020, (4): 158–163 |
|
||