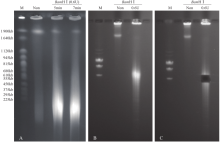
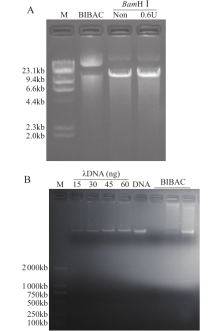
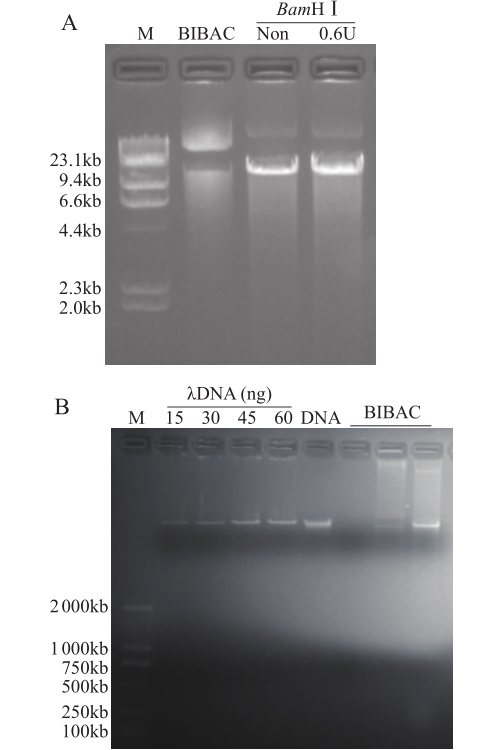
Crops ›› 2020, Vol. 36 ›› Issue (5): 103-109.doi: 10.16035/j.issn.1001-7283.2020.05.016
Previous Articles Next Articles
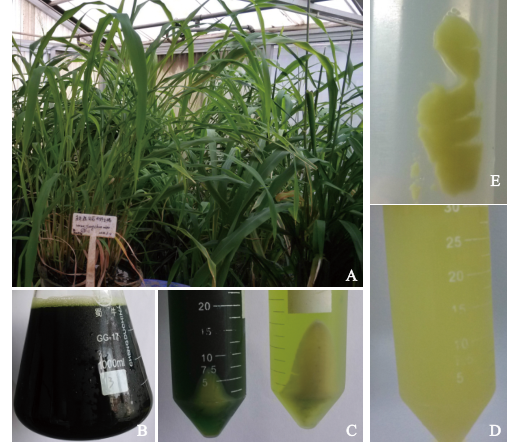
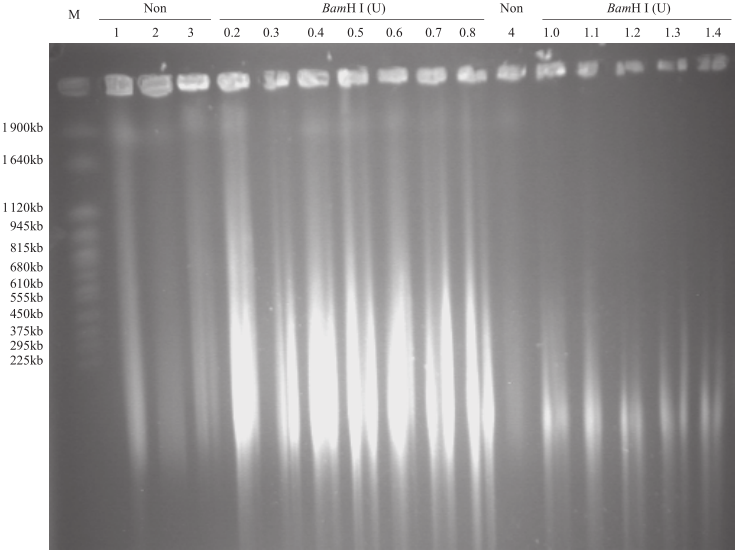
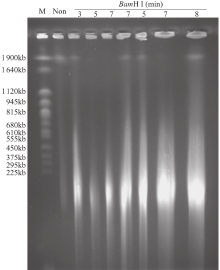
Extraction of High-Quality Chromosome DNA in Yunnan Wild Rice Oryza officinalis
Li Exian( ), Yin Fuyou, Zhang Dunyu, Chen Yue, Yu Tengqiong, Lei Yongtao, Xiao Suqin, Cheng Zaiquan, Ke Xue(
), Yin Fuyou, Zhang Dunyu, Chen Yue, Yu Tengqiong, Lei Yongtao, Xiao Suqin, Cheng Zaiquan, Ke Xue( )
)
- Institute of Biotechnology and Germplasm Resources, Yunnan Academy of Agricultural Sciences/Key Laboratory of Southwestern Crop Gene Resources and Germplasm Innovation, Ministry of Agriculture and Rural Affairs/Rice Materials Engineering Technology Research Center of Yunnan Province, Kunming 650205, Yunnan, China
| [1] | 程在全, 黄兴奇, 钱君, 等. 珍稀濒危植物——云南药用野生稻自然生态群的新发现及其特性. 云南植物研究, 2004,26(3):267-274. |
| [2] | 杨雅云, 张敦宇, 陈玲, 等. 云南药用野生稻对四种水稻主要病害的抗性鉴定. 植物病理学报, 2019,49(1):101-112. |
| [3] | 阿新祥, 吴成军, 鄢波, 等. 云南药用野生稻核基因组细菌人工染色体(BAC)文库的构建与分析. 植物生理学通讯, 2005,41(1):70-72. |
| [4] | 苏龙, 徐志健, 乔卫华 .等. 广西药用野生稻遗传多样性分析及SSR引物数量对遗传多样性结果的影响研究. 植物遗传资源学报, 2017,18(4):603-610. |
| [5] | 段峰森, 刘虹, 陈雁, 等. 利用C基因组DNA和C0t-1DNA对药用野生稻、大颖野生稻基因组的比较分析. 湖北农业科学, 2011,50(12):2548-2552. |
| [6] | 齐兰, 王效宁, 张吉贞, 等. 利用SRAP标记研究海南野生稻的遗传多样性与遗传分化. 植物遗传资源学报, 2013,14(3):402-406. |
| [7] | 吴绮, 覃瑞, 李刚, 等. 利用药用野生稻C基因组C0t-1 DNA比较分析宽叶野生稻CCDD基因组. 植物遗传资源学报, 2010,11(5):589-592. |
| [8] | 刘如亮, 刘虹, 陈雁, 等. 药用野生稻(CC)–栽培稻(AA)异源单体附加系MAAL8的遗传分析. 安徽农业科学, 2011,39(17):10139-10142. |
| [9] | 柯杰, 祝贺, 覃瑞, 等. 药用野生稻–栽培稻渗入系后代卷叶表型的遗传分析. 安徽农业科学, 2016,44(3):119-122. |
| [10] | 李培纲, 李红梅, 张帅, 等. 药用野生稻ADF基因的克隆及其抗逆性分析. 华北农学报, 2017,32(6):37-44. |
| [11] | 侯思名, 刘凌云, 殷旭东, 等. 药用野生稻BIBAC文库构建过程中质粒提取方法改进. 西南农业学报, 2014,27(1):1-5. |
| [12] | 张大燕, 李红梅, 李培纲, 等. 药用野生稻OoLEA1基因的克隆和生物信息学分析. 分子植物育种, 2017,15(5):1597-1602. |
| [13] | 汪暖, 陈志雄, 刘蕊, 等. 药用野生稻TAC克隆转化籼稻的体系初探. 植物生理学通讯, 2010,46(3):217-222. |
| [14] | 秦宗燕, 刘石娟, 包颖. 药用野生稻叶中花色苷合成基因的序列特点与表达差异. 植物生理学报, 2016,52(8):1248-1254. |
| [15] | 张欢欢, 刘蕊, 郭海滨, 等. 药用野生稻有利基因发掘与利用研究进展. 中国农学通报 2009,25(19):42-45. |
| [16] | 侯思名, 张薇, 白现广, 等. 野生稻细胞核DNA提取的研究. 西南农业学报, 2009,22(5):1215-1218. |
| [17] | 侯思名, 张薇, 翟书华, 等. 云南药用野生稻BIBAC文库的构建及分析. 湖南农业大学学报, 2011,37(1):17-21. |
| [18] | 刘艳平, 程在全, 殷富有, 等. 云南药用野生稻BIBAC文库混合克隆池制备及筛选. 生物技术通讯, 2012(2):117-120. |
| [19] | 王琳, 戴陆园, 吴丽华, 等. 云南三种野生稻原生境植物种群的调查及比较分析. 中国水稻科学, 2006,20(1):47-52. |
| [20] | 康公平, 徐国云, 陈志, 等. 茶陵普通野生稻光合特性研究. 作物学报, 2007,33(9):1558-1562. |
| [21] | 徐玲玲, 陈善娜, 程在全, 等. 云南野生稻离子吸收效率及其动力学特征研究. 植物生理学通讯, 2006,42(3):406-410. |
| [22] | 蒋春苗, 黄兴奇, 李定琴, 等. 云南野生稻叶茎根组织结构特性的比较研究. 西北植物学报, 2012,32(1):99-105. |
| [23] | Hamilton C M, Frary A, Lewis C, et al. Stable transfer of intact high molecular weight DNA into plant chromosomes. Proceedings of the National Academy of Sciences of the United States of America, 1996,93(18):9975-9979. |
| [24] |
Hamilton C M. A binary-BAC system for plant transformation with high-molecular-weight DNA. Gene, 1997,200(2):107-116.
doi: 10.1016/S0378-1119(97)00388-0 |
| [25] |
Hamilton C M, Frary A, Xu Y M, et al. Construction of tomato genomic DNA libraries in a binary-BAC (BIBAC) vector. Plant Journal, 1999,18(2):223-229.
doi: 10.1046/j.1365-313X.1999.00433.x |
| [26] |
Chang Y L, Chuang H W, Meksem K, et al. Characterization of a plant-transformation-ready large-insert BIBAC library of Arabidopsis and bombardment transformation of a large-insert BIBAC of the library into tobacco. Genome, 2011,54(6):437-447.
doi: 10.1139/G11-011 |
| [27] | Wang W Q, Wu Y R, Li Y, et al. A large insert Thellungiella halophila BIBAC library for genomics and identification of stress tolerance genes. Plant Molecular Biology, 2010,72(91):93-99. |
| [28] |
Fu Y H, Scheuring C F, Wang H Y, et al. Construction and characterization of a BIBAC library of Jatropha curcas L.and identification of BIBAC clones containing genes associated with fatty acid metabolism. Molecular Breeding, 2011,28(4):559-567.
doi: 10.1007/s11032-010-9505-2 |
| [29] |
Lee M, Zhang Y, Zhang M P, et al. Construction of a plant-transformation-competent BIBAC library and genome sequence analysis of polyploid upland cotton (Gossypium hirsutum L.). BMC Genomics, 2013,14:208-221.
doi: 10.1186/1471-2164-14-208 pmid: 23537070 |
| [30] | 成志伟, 曹筑荣, 陈良碧, 等. 东乡野生稻双元细菌人工染色体(BIBAC)文库的构建. 生物技术通报, 2006(zl):272-275,282. |
| [31] | 何瑞锋. 药用野生稻基因组文库构建与大片段DNA转化. 武汉:武汉大学, 2003. |
| [32] |
Thomas G, Joseph L, Varghese G, et al. Discrimination between Oryza malampuzhaensis Krish. et Chand. and Oryza officinalis Wall ex Watt based on RAPD markers and morphological traits. Euphytica, 2001,122(1):181-189.
doi: 10.1023/A:1012602516236 |
| [33] |
Naredo M E B, Mercado S M Q, Banaticla-Hilario M C N, et al. Genetic diversity patterns in ex situ collections of Oryza officinalis Wall. ex G. Watt revealed by morphological and microsatellite markers. Genetic Resources and Crop Evolution, 2017,64(4):733-744.
doi: 10.1007/s10722-016-0396-x |
| [34] | 闫影, 王春连, 刘丕庆, 等. 野生稻抗病虫基因的挖掘和利用. 作物杂志, 2011(4):15-19. |
| [35] | 陈明亮, 陈大洲, 胡兰香, 等. 东乡野生稻耐冷分子机制研究进展. 作物杂志, 2016(4):15-19. |
| [36] |
Tang H W, Zheng X M, Li C L, et al. Multi-step formation, evolution, and functionalization of new cytoplasmic male sterility genes in the plant mitochondrial genomes. Cell Research, 2017,27:130-146.
doi: 10.1038/cr.2016.115 pmid: 27725674 |
| [37] |
Mickelbart M V, Hasegawa P M, Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nature Reviews Genetics, 2015,16(4):237-251.
doi: 10.1038/nrg3901 pmid: 25752530 |
| [38] | 冯锐, 郭辉, 秦学毅, 等. 利用广西普通野生稻创新选育抗白叶枯病优质水稻三系不育系. 作物杂志, 2014(4):64-67. |
| [39] | Zou X H, Du Y S, Tang L, et al. Multiple origins of BBCC allopolyploid species in the rice genus (Oryza). Scientific Reports, 2015,5:1-8. |
| No related articles found! |
|
||