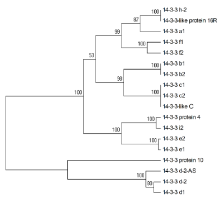
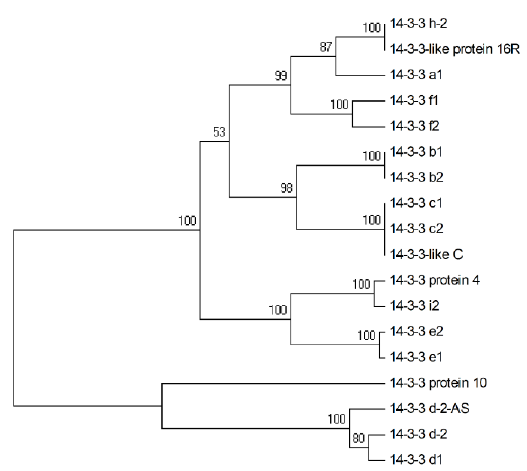
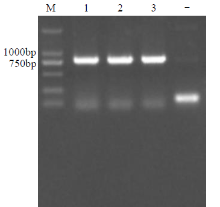
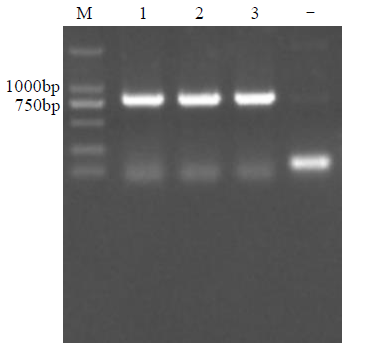
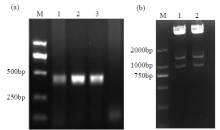
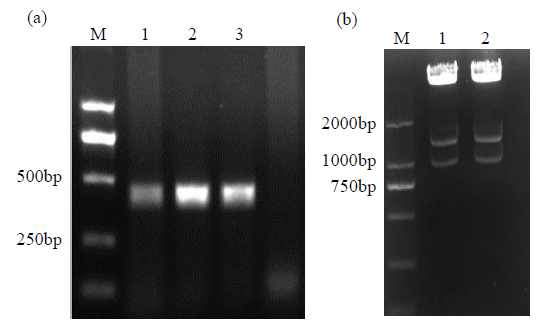
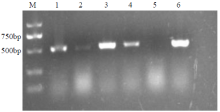
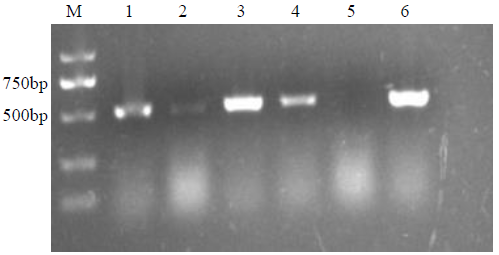
Crops ›› 2021, Vol. 37 ›› Issue (5): 6-13.doi: 10.16035/j.issn.1001-7283.2021.05.002
Previous Articles Next Articles
Cloning and Functional Analysis of Tobacco Nt14-3-3-like C Gene
Song Kunfeng1( ), Hao Fengsheng2, Yao Xin1, Wang Jing1, Liu Weiqun1(
), Hao Fengsheng2, Yao Xin1, Wang Jing1, Liu Weiqun1( )
)
- 1College of Life Sciences, Henan Agricultural University, Zhengzhou 450002, Henan, China
2College of Plant Protection, China Agricultural University, Beijing 100089, China
| [1] | 黄筱玲. 植物病毒病化学防治剂的研究现状及应用前景. 湖北化工, 1996, 13(S1):21-23. |
| [2] | 林祥永, 陈飞雄, 王金文, 等. 烟草花叶病的发生规律与防治策略. 安徽农业科学, 2003, 31(3):487. |
| [3] | 陈启建, 刘国坤, 吴祖建, 等. 三叶鬼针草中黄酮甙对烟草花叶病毒的抑制作用. 福建农林大学学报(自然科学版), 2003, 32(2):181-184. |
| [4] | 颜培强, 白先权, 万秀清, 等. 应用RNAi技术培育抗TMV病毒转基因烟草. 遗传, 2007, 29(8):1018-1022. |
| [5] |
Stange C, Matus J T, Elorza A, et al. Identification and characterization of a novel tobacco mosaic virus resistance N gene homologue in Nicotiana tabacum plants. Functional Plant Biology, 2004, 31(2):149-158.
doi: 10.1071/FP03160 |
| [6] |
Wang J, Wang X R, Qi Z, et al. iTRAQ protein profile analysis provides integrated insight into mechanisms of tolerance to TMV in tobacco (Nicotiana tobacum). Journal of Proteomics, 2016, 132(2):21-30.
doi: 10.1016/j.jprot.2015.11.009 |
| [7] |
Bachmann M, Huber J L, Liao P C, et al. The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14-3-3 protein. Federation of European Biochemical Societies Letters, 1996, 387(2/3):127.
doi: 10.1016/0014-5793(96)00478-4 |
| [8] |
Soll T M. 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. The Plant Cell, 2000, 12(1):53-64.
doi: 10.1105/tpc.12.1.53 |
| [9] |
Denison F C, Paul A L, Zupanska A K, et al. 14-3-3 proteins in plant physiology. Seminars in Cell and Developmental Biology, 2011, 22(7):720-727.
doi: 10.1016/j.semcdb.2011.08.006 pmid: 21907297 |
| [10] | Nina J, Christian T, Claudia O. Arabidopsis 14-3-3 proteins: fascinating and less fascinating aspects. Frontiers in Plant Science, 2011, 2(96):96. |
| [11] | Albertus H, Boer, Paula J, et al. Plant 14-3-3 proteins as spiders in a web of phosphorylation. Protoplasm A, 2013, 250(2):425-440. |
| [12] | Cotelle V, Leonhardt N. 14-3-3 proteins in guard cell signaling. Frontiers in Plant Science, 2016, 6:1210. |
| [13] |
Liu Q, Zhang S, Liu B. 14-3-3 proteins:macro-regulators with great potential for improving abiotic stress tolerance in plants. Biochemical and Biophysical Research Communications, 2016, 477(1):9-13.
doi: 10.1016/j.bbrc.2016.05.120 |
| [14] |
Taylor K W, Kim J G, Su X B, et al. Tomato TFT1 is required for PAMP-triggered immunity and mutations that prevent T3S effector xopN from binding to TFT1 attenuate xanthomonas virulence. PLoS Pathogens, 2012, 8(6):e1002768.
doi: 10.1371/journal.ppat.1002768 |
| [15] | Konagaya K I, Matsushita Y, Kasahara M, et al. Members of 14-3-3 protein isoforms interacting with the resistance gene product N and the elicitor of tobacco mosaic virus. Journal of General Plant Pathology, 2004, 70(4):221-231. |
| [16] | Malgorzata J R, Katarzyna K. The role of plasma membrane H+-ATPase in salinity stress of plants. Progress in Botany, 2015, 76:77-92. |
| [17] |
Viso F D, Casaretto J A, Quatrano R S. 14-3-3 proteins are components of the transcription complex of the ATEM1 promoter in Arabidopsis. Planta, 2007, 227(1):167-175.
doi: 10.1007/s00425-007-0604-1 |
| [18] |
周颖, 李冰樱, 李学宝. 14-3-3蛋白对植物发育的调控作用. 植物学报, 2012, 47(1):55-64.
doi: 10.3724/SP.J.1259.2012.00055 |
| [1] | Liu Xiaoli, Han Litao, Wei Nan, Shen Fei, Cai Yilin. Cloning, Molecular Characteristics Analysis and Genetic Transformation of Arabidopsis thaliana of ZmGS5 Gene in Maize [J]. Crops, 2021, 37(1): 16-25. |
| [2] | Haibin Luo, Shengli Jiang, Chengmei Huang, Huiqing Cao, Zhinian Deng, Kaichao Wu, Lin Xu, Zhen Lu, Yuanwen Wei. Cloning and Expression of ScHAK10 Gene in Sugarcane [J]. Crops, 2018, 34(4): 53-61. |
|
||