Crops ›› 2023, Vol. 39 ›› Issue (4): 22-30.doi: 10.16035/j.issn.1001-7283.2023.04.004
Previous Articles Next Articles
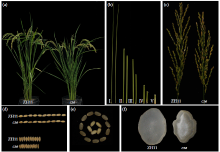
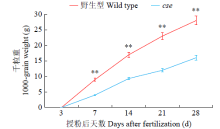
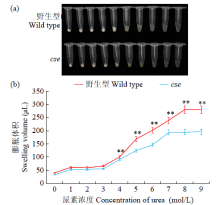
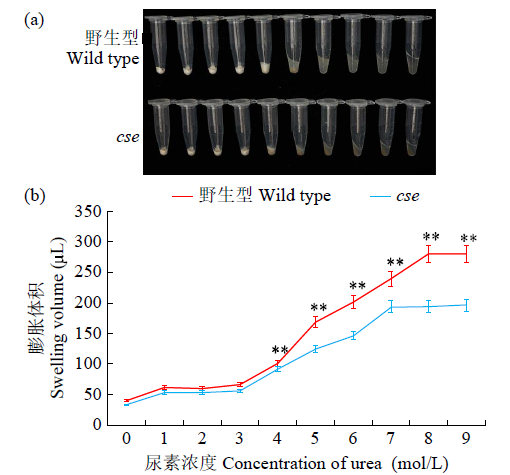
Phenotypic Analysis and Map-Based Cloning of Chalkiness and FlouryEndospermMutantcsein Rice (Oryza sativa L.)
Hua Qin1( ), Lin Quanxiang1, Song Yuanhui1, Sun Jiameng1, Zhang Zupu2,3, Chen Qingquan1, Li Jincai1, Zhang Haitao1(
), Lin Quanxiang1, Song Yuanhui1, Sun Jiameng1, Zhang Zupu2,3, Chen Qingquan1, Li Jincai1, Zhang Haitao1( )
)
- 1College of Agronomy, Anhui Agricultural University, Hefei230036, Anhui, China
2Jiangsu Hongqi Seed Co., Ltd., Taizhou225311, Jiangsu, China
3Jiangsu Province (Hongqi) Engineering Research Center for Rice and Wheat Seed Breeding, Taizhou225311, Jiangsu, China
| [1] | Wei X, Jiao G, Lin H, et al. GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development. Journal of Integrative Plant Biology, 2017, 59(2):134-153. |
| [2] | Hanashiro I, Itoh K, Kuratomi Y, et al. Granule-bound starch synthase I is responsible for biosynthesis of extra-long unit chains of amylopectin in rice. Plant Cell Physiology, 2008, 49(6):925- 933. |
| [3] | Dinges J R, Colleoni C, Myers A M, et al. Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiology, 2001, 125:1406-1418. |
| [4] | Zhou H, Wang L, Liu G, et al. Critical roles of soluble starch synthase SSIIIa and granule-bound starch synthase Waxy in synthesizing resistant starch in rice. Proceedings of the National Academy of Science of the United States of America, 2016, 113 (45):12844-12849. |
| [5] | Nayeon R, Chul Y, Cheon-Seok P, et al. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Reports, 2007, 26(7):1083- 1095. |
| [6] | She K C, Kusano H, Koizumi K, et al. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell, 2010, 22(10):3280-3294. |
| [7] | Kiswara G, Lee J H, Hur Y J, et al. Genetic analysis and molecular mapping of low amylose gene du12(t) in rice (Oryza sativa L.). Theoretical and Applied Genetics, 2014, 127(1):51-57. |
| [8] | Matsushima R, Maekawa M, Tomita K, et al. Amyloplast membrane protein SUBSTANDARD STARCH GRAIN6 controls starch grain size in rice endosperm. Plant Physiology, 2016, 170 (3):1445-1459. |
| [9] | Long W H, Wang Y H, Zhu S S, et al. FLOURY-SHRUNKEN ENDOSPERM1 connects phospholipid metabolism and amyloplast development in rice. Plant Physiology, 2018, 177(2):698-712. |
| [10] | Peng C, Wang Y H, Liu F, et al. FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant Journal, 2014, 77(6):917-930. |
| [11] | Lei J, Teng X, Wang Y F, et al. Plastidic pyruvate dehydrogenase complex E 1 component subunit Alpha1 is involved in galactolipid biosynthesis required for armyloplast development in rice. Plant Biotechnology Journal, 2022, 20(3):437-453. |
| [12] | Wang Y H, Liu F, Ren Y L, et al. GOLGI TRANSPORT 1B regulates protein export from endoplasmic reticulum in rice endosperm cells. Plant Cell, 2016, 28(11):2850-2865. |
| [13] | Ren Y L, Wang Y H, Pan T, et al. GPA5 encodes a Rab5a effector required for post-Golgi trafficking of rice storage proteins. Plant Cell, 2020, 32(3):758-777. |
| [14] | Zhu J P, Ren Y L, Wang Y L, et al. OsNHX5-mediated pH homeostasis is required for post-Golgi-trafficking of seed storage proteins in rice endosperm cells. BMC Plant Biology, 2019, 19 (1):295-307. |
| [15] | Wang J, Chen Z C, Zhang Q, et al. The NAC transcription factors OsNAC20 and OsNAC26 regulate starch and storage protein synthesis. Plant Physiology, 2020, 184(4):1775-1791. |
| [16] | Wu Y P, Pu C H, Lin H Y, et al. Three novel alleles of FLOURY ENDOSPERM2 (FLO2) confer dull grains with low amylose content in rice. Plant Science, 2015, 233:44-52. |
| [17] | Teramura H, Kondo K, Suzuki M, et al. Aberrant endosperm formation caused by reduced production of major allergen proteins in a rice flo2 mutant that confers low-protein accumulation in grains. Plant Biotechnology, 2019, 36(2):85-90. |
| [18] | 方鹏飞, 李三峰, 焦桂爱, 等. 水稻粉质胚乳突变体flo7的理化性质及基因定位. 中国水稻科学, 2014, 28(5):447-457. |
| [19] | Zhang L, Qi Y Z, Wu M M, et al. Mitochondrion-targeted PENTATRICOPEPTIDE REPEAT 5 is required for cis-splicing of nad4 intron 3 and endosperm development in rice. The Crop Journal, 2020, 9(2):282-296. |
| [20] | 中华人民共和国农业部. 米质测定方法:NY 147-88. 北京: 中国标准出版社. 2002. |
| [21] | 龙武华. 水稻粉质胚乳突变体flo8和fse的基因克隆及功能分析. 南京:南京农业大学, 2017. |
| [22] | 张述伟, 宗营杰, 方春燕, 等. 蒽酮比色法快速测定大麦叶片中可溶性糖含量的优化. 食品研究与开发, 2020, 41(7):196- 200. |
| [23] | 张斌. 稻米氨基酸含量的近红外定标模型的创建及应用. 杭州:浙江大学, 2010. |
| [24] | 赵玲珑. PTST2基因调控水稻花粉和胚乳淀粉积累的功能解析.扬州:扬州大学, 2020. |
| [25] | 于艳芳, 刘喜, 田云录, 等. 水稻粉质胚乳fse3突变体的表型分析及基因定位. 中国农业科学, 2018, 51(11):2023-2037. |
| [26] | Murray M G, Thompson W F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 1980, 8(19):4321- 4325. |
| [27] | Sanguinetti C J, Dias N E, Simpson A J. Rapid silver staining and recover of PCR products separated on polyacrylamide gels. Biotechniques, 1994, 17(5):915-919. |
| [28] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods, 2001, 25(4):402-408. |
| [29] | Kihira M, Tanicuchi K, Kaneko C, et al. Arabidopsis thaliana FLO2 is involved in efficiency of photoassimilate translocation, which is associated with leaf growth and aging, yield of seeds and seed quality. Plant Cell Physiology, 2017, 58(3):440-450. |
| [30] | Sato N, Kihira M, Matsushita R, et al. AtFLL2,a member of the FLO2 gene family, affects the enlargement of leaves at the vegetative stage and facilitates the regulation of carbon metabolism and flow. Bioscience Biotechnology and Biochemistry, 2020, 84 (12):2466-2475. |
| [31] | 张大鹏. 稻米品质突变体库的构建及粉质胚乳垩白突变体(flo6)的基因定位. 杭州:浙江大学, 2012. |
| [32] | Nishi A, Nakamura Y, Tanaka N, et al. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiology, 2001, 127(2):459-472. |
| [33] | Liabres S, Tsenkov M I, Macgowan S A, et al. Disease related single point mutations alter the global dynamics of a tetratricopeptide (TPR) α-solenoid domain. Journal of Structural Biology, 2020, 209(1):107405. |
| [34] | Wei G, Yang H X, Xiong Z X, et al. TPR domain coding gene ST2 may be involved in regulating tillering and fertility in rice. Czech Journal of Genetics and Plant Breeding, 2021, 57(3):83-90. |
| [35] | Myers A M, James M G, Lin Q, et al. Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell, 2011, 23(6):2331-2347. |
| [36] | Kelly A A, Kalisch B, Hölzl G, et al. Synthesis and transfer of galactolipids in the chloroplast envelope membranes of Arabidopsis thaliana. Proceedings of the National Academy Science of the United States of America, 2016, 113(38):10714- 10719. |
| [1] | Bingshu He,Yuyue Zhong,Yongli Qiao,Dongwei Guo. Analysis of Grain Traits and Physicochemical Properties of Starch in Rice Floury Endosperm Mutant flo(t) [J]. Crops, 2017, 33(4): 67-71. |
|
||