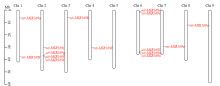
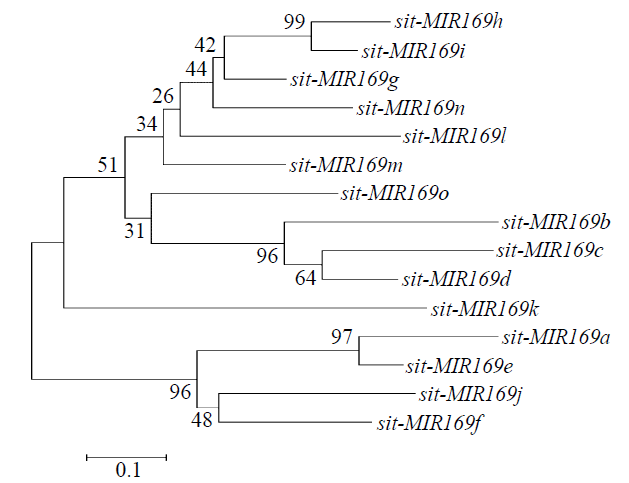
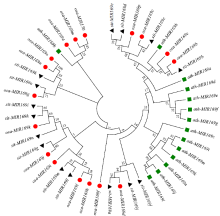
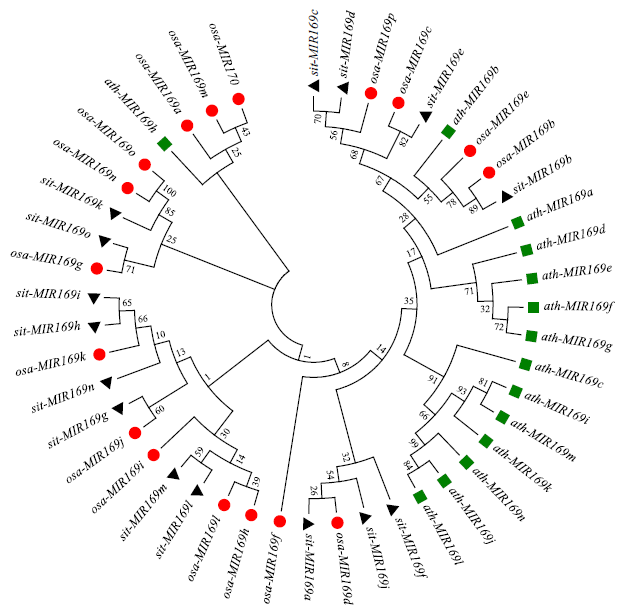
Crops ›› 2022, Vol. 38 ›› Issue (2): 54-63.doi: 10.16035/j.issn.1001-7283.2022.02.008
Previous Articles Next Articles
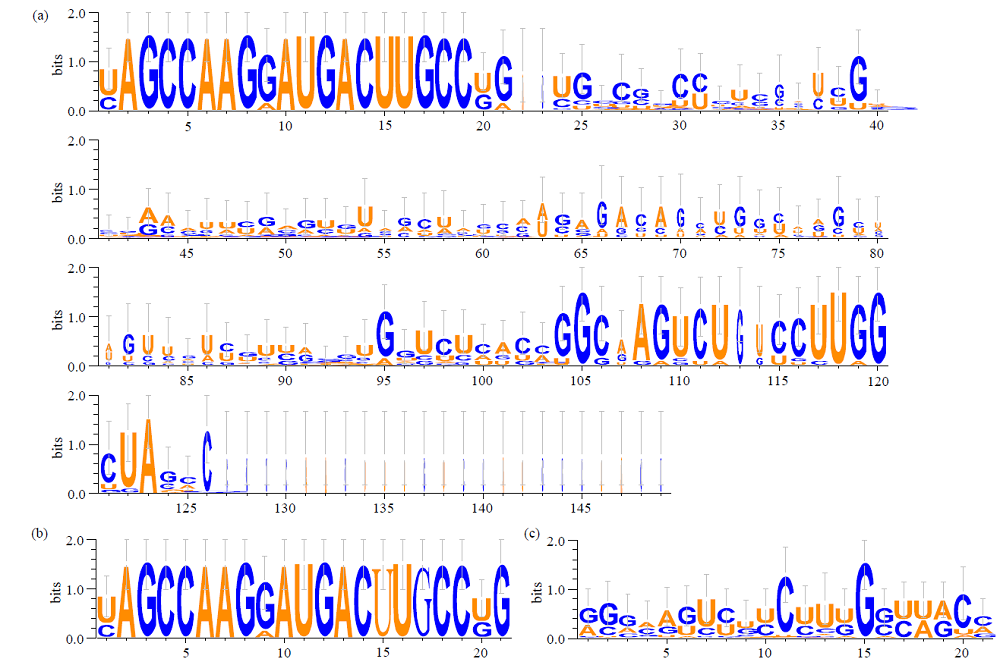
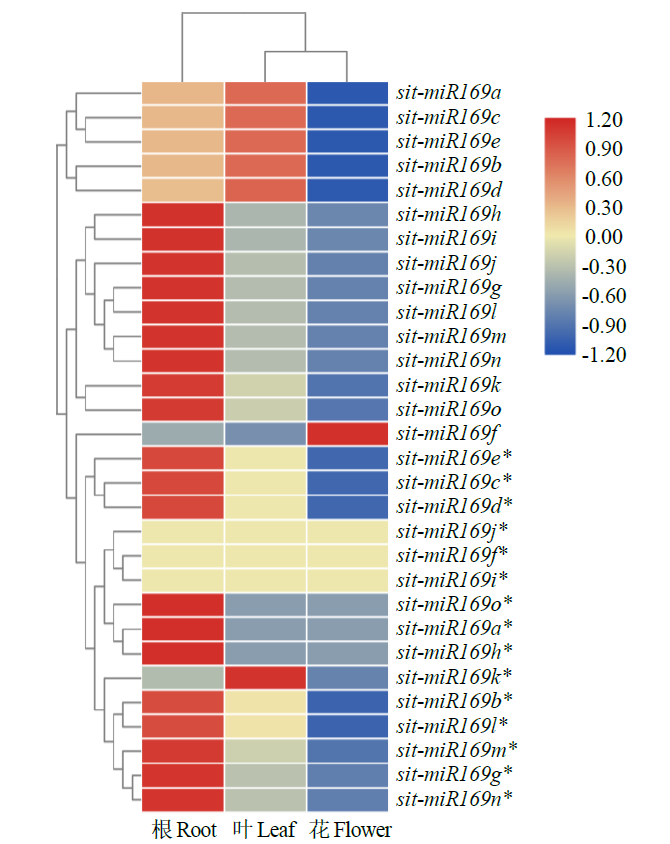
Identification and Functional Analysis of miR169 Family and Its Target Genes in Setaria italica
Lu Ping1( ), Kang Qingfang1, Zhao Mengyao1, Zhang Fengjie2, Wu Qiangqiang2, Ma Fangfang2, Wang Yushen2, Han Yuanhuai2, Wang Xingchun1, Li Xueyin1,*(
), Kang Qingfang1, Zhao Mengyao1, Zhang Fengjie2, Wu Qiangqiang2, Ma Fangfang2, Wang Yushen2, Han Yuanhuai2, Wang Xingchun1, Li Xueyin1,*( )
)
- 1College of Life Sciences, Shanxi Agricultural University, Jinzhong 030801, Shanxi, China
2College of Agriculture, Shanxi Agricultural University, Jinzhong 030801, Shanxi, China
| [1] | 刁现民. 中国谷子产业与产业技术体系. 北京: 中国农业科学技术出版社, 2011. |
| [2] |
Diao X M, Schnable J, Bennetzen J L, et al. Initiation of Setaria as a model plant. Frontiers of Agricultural Science and Engineering, 2014, 1(1):16.
doi: 10.15302/J-FASE-2014011 |
| [3] |
Yang Z R, Zhang H S, Li X K, et al. A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nature Plants, 2020, 6(9):1167-1178.
doi: 10.1038/s41477-020-0747-7 |
| [4] |
Bennetzen J L, Schmutz J, Wang H, et al. Reference genome sequence of the model plant Setaria. Nature Biotechnology, 2012, 30(6):555-561.
doi: 10.1038/nbt.2196 pmid: 22580951 |
| [5] |
Zhang G Y, Liu X, Quan Z W, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nature Biotechnology, 2012, 30(6):549-554.
doi: 10.1038/nbt.2195 |
| [6] |
杨丽娟, 李世访, 卢美光. miRNA 在植物病原调控方面的研究进展. 生物技术通报, 2020, 36(1):101-109.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-1045 |
| [7] | 郁佳雯, 郁佳雯, 韩荣鹏, 等. microRNA 在植物生长发育中的研究进展. 分子植物育种, 2020, 18(5):1496-1504. |
| [8] |
张翠桔, 莫蓓莘, 陈雪梅, 等. 植物 miRNA 作用方式的分子机制研究进展. 生物技术通报, 2020, 36(7):1-14.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0262 |
| [9] | Wang Y Q, Li L, Tang S, et al. Combined small RNA and degradome sequencing to identify miRNAs and their targets in response to drought in foxtail millet. BMC Genetics, 2016, 17(1):1-16 |
| [10] |
Xu M Y, Zhang L, Li W W, et al. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. Journal of Experimental Botany, 2014, 65(1):89-101.
doi: 10.1093/jxb/ert353 |
| [11] | 赵胜利.Osa-miR169 调控水稻对稻瘟病菌免疫机理的研究. 成都:四川农业大学, 2017. |
| [12] |
Zhao B T, Ge L F, Liang R Q, et al. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Molecular Biology, 2009, 10(1):1-10.
doi: 10.1186/1471-2199-10-1 |
| [13] |
Luan M D, Xu M Y, Lu Y M, et al. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene, 2015, 555(2):178-185.
doi: 10.1016/j.gene.2014.11.001 |
| [14] |
Ni Z Y, Hu Z, Jiang Q Y, et al. GmNFYA3,a target gene of miR169,is a positive regulator of plant tolerance to drought stress. Plant Molecular Biology, 2013, 82(1/2):113-129.
doi: 10.1007/s11103-013-0040-5 |
| [15] | 尹海龙.大豆 miR169d 的表达分析及功能验证. 长春:吉林农业大学, 2013. |
| [16] | 孙广鑫, 栾雨时, 崔娟娟. 番茄中与致病密切相关 miRNA 的挖掘及特性分析. 遗传, 2013, 36(1):69-76. |
| [17] | 董云, 王毅, 靳丰蔚, 等. 油菜 Bna-miR169d 基因的分离与过表达初步分析. 西北农业学报, 2016, 25(12):1809-1815. |
| [18] | 刘志祥, 曾超珍, 谭晓风. 杨树 MIR169 基因家族分子进化分析. 遗传, 2013, 35(11):1307-1316. |
| [19] | 方辉, 曲俊杰, 孙嘉曼, 等. 葡萄 miR169 及其靶基因的生物信息学分析. 南方农业学报, 2017, 48(8):1329-1334. |
| [20] | Donati G, Gatta R, Dolfini D, et al. An NF-Y-dependent switch of positive and negative histone methyl marks on CCAAT promoters. PLoS ONE, 2008, 3(4):e2066. |
| [21] | Nelson D E, Repetti P P, Adams T R, et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proceedings of the National Academy of Sciences, 2007, 104(42):16450-16455. |
| [22] | Sinha S, Maity S N, Lu J F, et al. Recombinant rat CBF-C,the third subunit of CBF/NFY,allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proceedings of the National Academy of Sciences, 1995, 92(5):1624-1628. |
| [23] |
Li W X, Oono Y, Zhu J H, et al. The Arabidopsis NFYA 5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. The Plant Cell, 2008, 20(8):2238-2251.
doi: 10.1105/tpc.108.059444 |
| [24] | Guo Z L, Kuang Z, Wang Y, et al. PmiREN:a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Research, 2020, 48(D1):1114-1121. |
| [25] |
Chen C J, Chen H, Zhang Y, et al. TBtools-an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 2020, 13(8):1194-1202.
doi: 10.1016/j.molp.2020.06.009 |
| [26] |
Crooks G E, Hon G, Chandonia J M, et al. WebLogo:a sequence logo generator. Genome Research, 2004, 14(6):1188-1190.
doi: 10.1101/gr.849004 pmid: 15173120 |
| [27] | Dai X B, Zhuang Z H, Zhao P X. psRNATarget:a plant small RNA target analysis server (2017 release). Nucleic Acids Research, 2018, 46(W1):49-54. |
| [28] |
Zhang Z H, Teotia S, Tang J H, et al. Perspectives on microRNAs and phased small interfering RNAs in maize (Zea mays L.):functions and big impact on agronomic traits enhancement. Plants, 2019, 8(6):1-17.
doi: 10.3390/plants8010001 |
| [29] |
Devos K M, Wang Z M, Beales J, et al. Comparative genetic maps of foxtail millet (Setaria italica) and rice (Oryza sativa). Theoretical and Applied Genetics, 1998, 96(1):63-68.
doi: 10.1007/s001220050709 |
| [30] | 徐妙云, 朱佳旭, 张敏, 等. 植物miR169/NF-YA调控模块研究进展. 遗传, 2016, 38(8):700-706. |
| [31] | 戴晓峰, 肖玲, 武玉花, 等. 植物脂肪酸去饱和酶及其编码基因研究进展. 植物学报, 2007, 24(1):105-113. |
| [1] | Yin Guifang, Duan Ying, Yang Xiaolin, Cai Suyun, Wang Yanqing, Lu Wenjie, Sun Daowang, He Runli, Wang Lihua. Cloning and Bioinformatics Analysis of FtC4H Gene from Tartary Buckwheat [J]. Crops, 2022, 38(1): 77-83. |
| [2] | Zhao Xunchao,Xu Jingyu,Gai Shengnan,Wei Yulei,Xu Xiaoxuan,Ding Dong,Liu Meng,Zhang Jinjie,Shao Wenjing. Identification of Stearyl -ACP Desaturase Gene (SbSAD) Family and Their Expression Analysis at Different Developmental Stages in Sorghum [J]. Crops, 2020, 36(2): 20-27. |
| [3] | Song Jian,Cao Xiaoning,Wang Haigang,Chen Ling,Wang Junjie,Liu Sichen,Qiao Zhijun. Identification and Expression Analysis of ASR Family Genes in Setaria italica [J]. Crops, 2019, 35(6): 33-42. |
| [4] | Yue Linqi,Shi Weiping,Guo Jiahui,Guo Pingyi,Guo Jie. Response of Cutin Synthetic Genes of Foxtail Millet to Drought Stress [J]. Crops, 2019, 35(4): 183-190. |
| [5] | Lü Liangjie,Chen Xiyong,Zhang Yelun,Liu Qian,Wang Limei,Ma Le,Li Hui. Bioinformatics Identification of GASA Gene Family Expression Profiles in Wheat [J]. Crops, 2018, 34(6): 58-67. |
| [6] | Haibin Luo, Shengli Jiang, Chengmei Huang, Huiqing Cao, Zhinian Deng, Kaichao Wu, Lin Xu, Zhen Lu, Yuanwen Wei. Cloning and Expression of ScHAK10 Gene in Sugarcane [J]. Crops, 2018, 34(4): 53-61. |
| [7] | Ying Zhang,Pengyu Liu,Xue Bai,Yang Yang,Yueying Li. Expression and Bioinformatics Analysis of CsWRKY23 Gene in Cucumber [J]. Crops, 2017, 33(5): 38-42. |
|
||