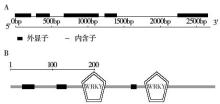
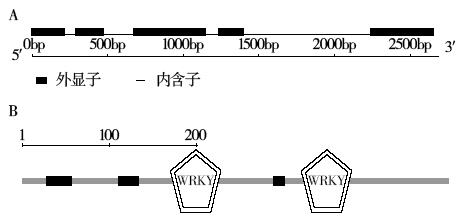
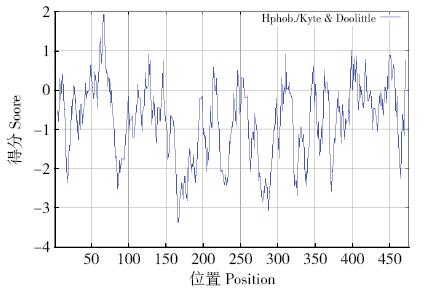
Crops ›› 2017, Vol. 33 ›› Issue (5): 38-42.doi: 10.16035/j.issn.1001-7283.2017.05.007
Previous Articles Next Articles
Expression and Bioinformatics Analysis of CsWRKY23 Gene in Cucumber
Zhang Ying1,Liu Pengyu2,Bai Xue1,Yang Yang1,Li Yueying1
- 1College of Life Science,Shenyang Normal University,Shenyang 110034,Liaoning,China
2Shenyang Institute of Food Control,Shenyang 110136,Liaoning,China
| [1] |
Rushton P J, Somssich I E, Ringler P , et al. WRKY transcription factors. Trends in Plant Science 2010,15(5):247-258.
doi: 10.1016/j.tplants.2010.02.006 |
| [2] |
张颖, 蒋卫杰, 凌键 , 等. WRKY转录因子表达谱的研究进展. 基因组学与应用生物学, 2009,28(4):1-6.
doi: 10.3969/gab.028.000803 |
| [3] |
陈思雀, 翁群清, 曹红瑞 , 等. WRKY转录因子在生物和非生物胁迫中的功能和调控机理的研究进展. 农业生物技术学报, 2017,25(4):668-682.
doi: 10.3969/j.issn.1674-7968.2017.04.017 |
| [4] |
Samad A F A, Sajad M, Nazaruddin N , et al. MicroRNA and transcription factor:key players in plant regulatory network. Frontiers in Plant Science, 2017,8:565.
doi: 10.3389/fpls.2017.00565 pmid: 5388764 |
| [5] |
Huang S, Li R, Zhang Z , et al. The genome of the cucumber,Cucumis sativus L. Nature Genetics, 2009,41:1275-1281.
doi: 10.1038/ng.475 pmid: 19881527 |
| [6] |
Zhu J, Dong C H, Zhu J K . Interplay between cold-responsive gene regulation,metabolism and RNA processing during plant cold acclimation. Current Opinion in Plant Biology, 2007,10(3):290-295.
doi: 10.1016/j.pbi.2007.04.010 |
| [7] |
Livak K J, Schmittgen T D . Analysis of relative gene expression data using real-time quantitative PCR and the 2 -△△CT method . Methods, 2001,25(4):402-408.
doi: 10.1006/meth.2001.1262 |
| [8] |
Eulgem T, Rushton P J, Robatzek S , et al. The WRKY superfamily of plant transcription factors. Trends in Plant Science, 2000,5(5):199-205.
doi: 10.1016/S1360-1385(00)01600-9 pmid: 10785665 |
| [9] |
Ling J, Jiang W, Zhang Y , et al. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC genomics, 2011,12(1):471.
doi: 10.1186/1471-2164-12-471 pmid: 3191544 |
| [10] |
Jiang J, Ma S, Ye N , et al. WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology, 2017,59(2):86-101.
doi: 10.1111/jipb.12513 pmid: 27995748 |
| [11] | 喻景权, 周杰 . “十二五”我国设施蔬菜生产和科技进展及其展望.中国蔬菜, 2016(9):18-30. |
| [12] |
陈明亮, 陈大洲, 胡兰香 , 等. 东乡野生稻耐冷分子机制研究进展.作物杂志, 2016(4):15-19.
doi: 10.16035/j.issn.1001-7283.2016.04.003 |
| [13] |
秦东玲, 李钊, 尉菊萍 , 等. 作物抗冷性及其化学控制机理研究进展.作物杂志, 2016(4):26-35.
doi: 10.16035/j.issn.1001-7283.2016.04.005 |
| [14] |
Tripathi P, Rabara R C, Rushton P J . A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta, 2014,239(2):255-266.
doi: 10.1007/s00425-013-1985-y |
| [15] | Kim C Y , Vo K T X,Nguyen C D,et al.Functional analysis of a cold-responsive rice WRKY gene,OsWRKY71. Plant Biotechnology Reports, 2016,10(1):12-23. |
| [16] | 魏小春, 姚秋菊, 原玉香 , 等. 辣椒CaWRKY13基因克隆及非生物胁迫下表达分析. 分子植物育种, 2016,14(10):2582-2588. |
| [17] |
Zhang Y, Yu H, Yang X , et al. CsWRKY46,a WRKY transcription factor from cucumber,confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiology and Biochemistry, 2016,108:478-487.
doi: 10.1016/j.plaphy.2016.08.013 |
| [18] |
付乾堂, 余迪求 . 拟南芥AtWRKY25、AtWRKY26和AtWRKY33在非生物胁迫条件下的表达分析. 遗传, 2010,32(8):848-856.
doi: 10.3724/SP.J.1005.2010.00848 |
| [19] |
Zheng Z, Qamar S A, Chen Z , et al. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant Journal, 2006,48(4):592-605.
doi: 10.1111/tpj.2006.48.issue-4 |
| [20] |
Zhou J, Wang J, Zheng Z Y , et al. Characterization of the promoter and extended C-terminal domain of Arabidopsis WRKY33 and functional analysis of tomato WRKY33 homologues in plant stress responses. Journal of Experimental Botany, 2015,66(15):4567-4583.
doi: 10.1093/jxb/erv221 |
| [1] | Haibin Luo, Shengli Jiang, Chengmei Huang, Huiqing Cao, Zhinian Deng, Kaichao Wu, Lin Xu, Zhen Lu, Yuanwen Wei. Cloning and Expression of ScHAK10 Gene in Sugarcane [J]. Crops, 2018, 34(4): 53-61. |
| [2] | Zhiwei Zhang,Xiaojing Li,Jinrui Bai,shuai Chen. Effects of CO2 on Cucumber Leaf Photosynthetic Rate and Stomatal Characters under High Temperature [J]. Crops, 2016, 32(5): 81-86. |
|