Crops ›› 2022, Vol. 38 ›› Issue (1): 31-37.doi: 10.16035/j.issn.1001-7283.2022.01.004
Previous Articles Next Articles
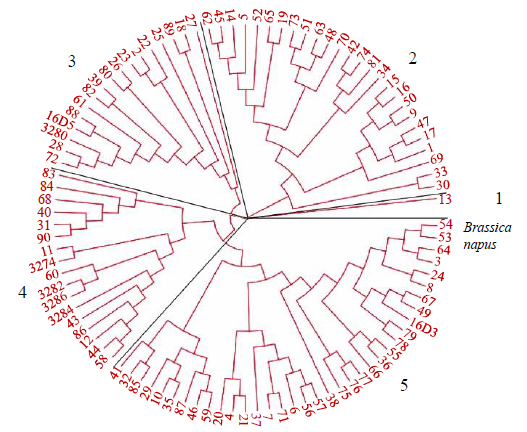
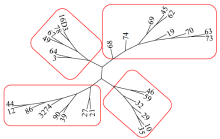
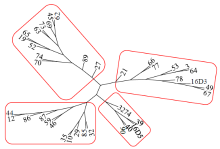
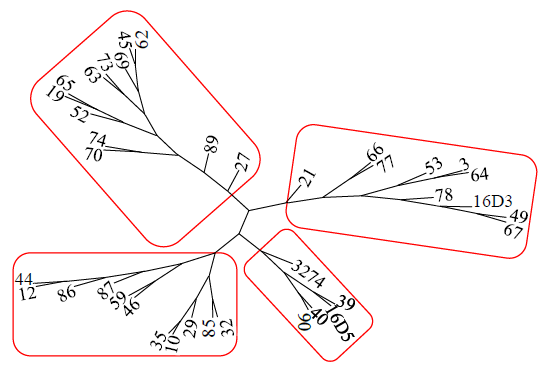
Core Collection Construction of Extra-Early Restorer Lines in Spring Brassica napus L.
Lin Xiaoyang( ), Du Dezhi, Liu Haidong(
), Du Dezhi, Liu Haidong( ), Li Jun
), Li Jun
- Academy of Agricultural and Forestry Sciences of Qinghai University/Key Laboratory of Spring Rapeseed Genetic Improvement/The Qinghai Research Branch of the National Rapeseed Genetic Improvement Center/ Spring Rape Scientific Observation Experimental Station of Ministry of Agriculture and Rural Affairs/Qinghai Research and Development Center for Spring Rapeseed/Qinghai Province Spring Rape Engineering Technology Research Center, Xining 810016, Qinghai, China
| [1] | 张亚妮, 姚艳梅. 118份春性甘蓝型油菜恢复系的桔红花色位点鉴定. 分子植物育种, 2020, 18(6):1893-1900. |
| [2] | 尹中江, 关卫星, 杨勇, 等. 育种平台(软件)在西藏作物育种中应用和西藏育种方向浅析. 西藏农业科技, 2020, 42(2):46-51. |
| [3] | 张鹤. 油菜表型精准鉴定及优良种质筛选. 杨凌:西北农林科技大学, 2019. |
| [4] | 潘云龙, 柳海东. 甘蓝型春油菜早花位点cqDTFA7a加密及其近等基因系构建. 分子植物育种, 2019, 17(21):7047-7057. |
| [5] | 姚艳梅, 徐亮, 胡琼, 等. 青海省几个主要春性甘蓝型油菜杂交种的SSR分析. 中国油料作物学报, 2008, 30(4):406-410. |
| [6] | 傅廷栋, 杨小牛. 甘蓝型油菜波里马雄性不育系的选育与研究. 华中农业大学学报, 1989, 8(3):7. |
| [7] | 丁厚栋, 张尧锋, 余华胜, 等. 甘蓝型油菜种质资源的农艺性状聚类分析. 华北农学报, 2009, 24(S1):103-105. |
| [8] | 李自超, 张洪亮, 曹永生, 等. 中国地方稻种资源初级核心种质取样策略研究. 作物学报, 2003, 29(1):20-24. |
| [9] | 王丽侠, 李英慧, 李伟, 等. 长江春大豆核心种质构建及分析. 生物多样性, 2004(6):578-585. |
| [10] | 郝晨阳, 董玉琛, 王兰芬, 等. 我国普通小麦核心种质的构建及遗传多样性分析. 科学通报, 2008(8):908-915. |
| [11] | 李强, 马代夫, 刘庆昌, 等. 中国北方薯区甘薯育种核心亲本初步构建与利用. 西北农业学报, 2010, 19(12):48-52. |
| [12] | 曾晓珊, 彭丹, 石媛媛, 等. 利用SSR标记构建水稻核心亲本指纹图谱. 作物研究, 2016, 30(5):481-486,511. |
| [13] | 杨勇. 甘蓝型油菜遗传多样性分析及核心亲本的指纹图谱构建. 武汉:华中农业大学, 2013. |
| [14] | Schoen D J, Brown A H. Conservation of allelic richness in wild crop relatives is aided by assessment of genetic markers. Proceedings of the National Academy of Sciences of the United States of America, 1993, 90(22):10623-10627. |
| [15] |
Chris T, José C, Jorge F, et al. Core Hunter:an algorithm for sampling genetic resources based on multiple genetic measures. BMC Bioinformatics, 2009, 10(1):243.
doi: 10.1186/1471-2105-10-243 |
| [16] | 杜德志, 肖麓, 赵志, 等. 我国春油菜遗传育种研究进展. 中国油料作物学报, 2018, 40(5):633-639. |
| [17] |
Kozich J J, Westcott S L, Baxter N T, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeqIllumina sequencing platform. Applied and Environmental Microbiology, 2013, 79(17):5112-5120.
doi: 10.1128/AEM.01043-13 pmid: 23793624 |
| [18] |
Li R, Yu C, Li Y, et al. SOAP2:an improved ultrafast tool for short read alignment. Bioinformatics, 2009, 25(15):1966-1967.
doi: 10.1093/bioinformatics/btp336 |
| [19] |
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 2009, 25(14):1754-1760.
doi: 10.1093/bioinformatics/btp324 |
| [20] |
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit:a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 2010, 20(9):1297-1303.
doi: 10.1101/gr.107524.110 pmid: 20644199 |
| [21] |
Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics, 2009, 25(16):2078-2079.
doi: 10.1093/bioinformatics/btp352 |
| [22] | 唐晓敏, 张春荣, 周良云, 等. 基于SLAF-Seq技术的广金钱草SNP位点开发及遗传分析. 分子植物育种, 2020, 18(18):6101-6107. |
| [23] | 赵绪涛, 柳海东, 李开祥, 等. 基于SLAF-seq技术构建甘蓝型油菜Polima CMS恢复系核心种质. 中国油料作物学报, 2019, 41(4):497-506. |
| [24] |
Kumar S, Glen S, Li M, et al. MEGA X:molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 2018, 35(6):1547-1549.
doi: 10.1093/molbev/msy096 |
| [25] | 邓念丹, 刘清波, 蒋建雄, 等. 五节芒核心种质的构建研究概述. 作物研究, 2010, 24(2):130-134. |
| [26] | 陈敬锋, 马金霞. 分子水平建议核心种质的算法初探. 新疆农业大学学报, 2001(2):26-29. |
| [27] | 何余堂, 涂金星, 傅廷栋, 等. 陕西省白菜型油菜核心种质的初步构建. 中国油料作物学报, 2002(1):7-10. |
| [28] | 姚艳梅, 唐国永. 特早熟春性甘蓝型油菜青杂8号的选育. 青海大学学报(自然科学版), 2015, 33(6):13-17,54. |
| [29] | 管传禧. 甘蓝型油菜早熟育种的体会. 安徽农业科学, 1984(1):46-49. |
| [30] | 杜德志, 李秀萍, 余青兰, 等. 特早熟双低甘蓝型油菜杂交种的选育. 西北农业学报, 2003(1):1-4. |
| [1] | Lü Weisheng, Xiao Xiaojun, Huang Tianbao, Xiao Guobin, Li Yazhen, Xiao Fuliang, Han Depeng, Zheng Wei. Application Effect of Slow-Released Formulated Fertilizer on Oilseed Rape (Brassica napus L.) under Late Sowing Rice [J]. Crops, 2020, 36(6): 143-150. |
|
||