Crops ›› 2022, Vol. 38 ›› Issue (3): 9-19.doi: 10.16035/j.issn.1001-7283.2022.03.002
Previous Articles Next Articles
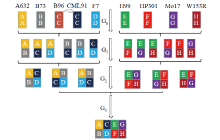
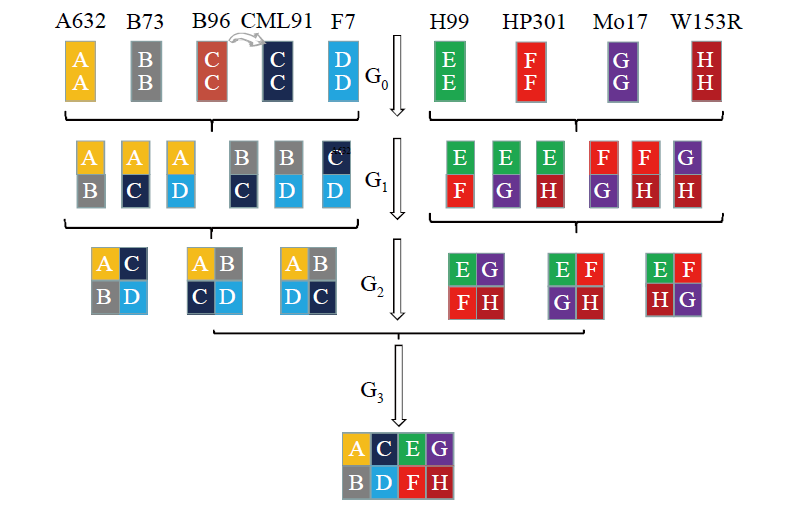
Genetic Characteristics of MAGIC Population and Its Application in Crop Stress Tolerance
Rong Kewei1,2( ), Liu Bojuan2, Lu Yuelei1,2, Chen Yong2, Luo Ping1,2, Zhao Kang1, Hao Zhuanfang1,2(
), Liu Bojuan2, Lu Yuelei1,2, Chen Yong2, Luo Ping1,2, Zhao Kang1, Hao Zhuanfang1,2( ), Gao Wenwei1(
), Gao Wenwei1( )
)
- 1College of Agronomy, Xinjiang Agricultural University, Urumqi 830052, Xinjiang, China
2Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
| [1] |
卢宝荣, 朱有勇, 王云月. 农作物遗传多样性农家保护的现状及前景. 生物多样性, 2002(4):409-415.
doi: 10.17520/biods.2002056 |
| [2] | 李晓方. 农作物多基因型种群育种及种子生产技术体系. 遗传, 2012, 34(3):382. |
| [3] | 罗开, 周永明, 张椿雨, 等. 甘蓝型油菜MAGIC群体的遗传结构分析及应用. 武汉:华中农业大学, 2015. |
| [4] |
Thornsberry J M, Goodman M M, Doebley J, et al. Dwarf8 polymorphisms associate with variation in flowering time. Nature Genetics, 2001, 28(3):286-289.
pmid: 11431702 |
| [5] | Bentsink L, Hanson J, Coltrane C, et al. Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(9):4264-4269. |
| [6] |
Ballini E, Morel J B, Droc G, et al. A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Molecular Plant-microbe Interactions:MPMI, 2008, 21(7):859-868.
doi: 10.1094/MPMI-21-7-0859 |
| [7] |
Yu J M, Holland J B, McMullen M D, et al. Genetic design and statistical power of nested association mapping in maize. Genetics, 2008, 178(1):539-551.
doi: 10.1534/genetics.107.074245 |
| [8] | Remington D L, Thornsberry J M, Matsuoka Y, et al. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proceedings of the National Academy of Sciences of the United States of America, 2001, 98:11479-11484. |
| [9] |
Bandillo N, Raghavan C, Muyco P A, et al. Multi-parent advanced generation inter-cross (MAGIC) populations in rice:progress and potential for genetics research and breeding. Rice, 2013, 6(1):1-15.
doi: 10.1186/1939-8433-6-1 |
| [10] |
Mason A S, Batley J, Bayer P E, et al. High-resolution molecular karyotyping uncovers pairing between ancestrally related Brassica chromosomes. The New phytologis, 2014, 202(3):964-974.
doi: 10.1111/nph.12706 |
| [11] |
Rakshit S, Rakshit A, Patil J V. Multiparent intercross populations in analysis of quantitative traits. Journal of Genetics, 2012, 91(1):111-117.
doi: 10.1007/s12041-012-0144-8 pmid: 22546834 |
| [12] | Cavanagh C, Morell M, Macky I, et al. From mutations to MAGIC:resources for gene discovery,validation and delivery in crop plants. Current Opinion in Plant Biology, 2007, 11(2):2-3. |
| [13] | Cavanagh C R, Chao S M, Wang S C, et al. Genome-wide comparative diversity uncovers multiple targets of selection for inprovement in hexaploid wheat landraces and cultivars. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(20):8057-8062. |
| [14] | Huang X Q, Paulo M J, Boer M, et al. Analysis of natural allelic variation in Arabidopsis using a multiparent recombinant inbred line population. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(11):4488-4493. |
| [15] |
Churchill G A, Airey D C, Allayee H, et al. The Collaborative Cross,a community resource for the genetic analysis of complex traits. Nature Genetics. 2004, 36(11):1133-1137.
pmid: 15514660 |
| [16] |
Huang X H, Wei X H, Zhao Q, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nature Genetics, 2010, 42(11):961-967.
doi: 10.1038/ng.695 |
| [17] | Mott R, Talbot C J, Turri M G, et al. A method for fine mapping quantitative trait loci in outbredanimal stocks. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(23):12649-12654. |
| [18] | Smallwood T L, Gatti D M, Weinstock G M, et al. High-resolution genetic mapping in the diversity outbred mouse population identifies Apobec1 as a candidate gene for atherosclerosis. G3:Genes, Genomes,Genetics, 2014, 4(12):2353-2363. |
| [19] | King E G, Merkes C M, McNeil C L, et al. Genetic dissection of a model complex trait using the Drosophila Synthetic population resource. Cold Spring Harbor Laboratory Press, 2012, 22(8):1558-1566. |
| [20] |
Macdonald S J, Long A D. Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics, 2007, 176(2):1261-1281.
pmid: 17435224 |
| [21] |
Marriage T N, King E G, Long A D, et al. Fine-mapping nicotine resistance loci in Drosophila using a multiparent advanced generation inter-cross population. Genetics, 2014, 198(1):45-57.
doi: 10.1534/genetics.114.162107 pmid: 25236448 |
| [22] | 杨媚. 水稻MAGIC群体的分子基础及生态响应的特性. 广州:华南师范大学, 2012. |
| [23] |
Meng L J, Zhao X Q, Ponce K, et al. QTL mapping for agronomic traits using Multi-parent Advanced Generation Inter-Cross (MAGIC) populations derived from diverse elite Indica rice lines. Field Crops Research, 2016, 189:19-42.
doi: 10.1016/j.fcr.2016.02.004 |
| [24] |
Broman K W. Genotype Probabilities at intermediate generations in the construction of recombinant inbred lines. Genetics, 2012, 190(2):403-412.
doi: 10.1534/genetics.111.132647 pmid: 22345609 |
| [25] | 胡刚. 利用水稻4亲本MAGIC群体进行粒形和株型的遗传分析. 武汉:华中农业大学, 2017. |
| [26] | Kover P X, Valdar W, Trakalo J, et al. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genetics, 2009, 5(7):e1000551. |
| [27] |
Xiao Y J, Jiang S Q, Cheng Q, et al. The genetic mechanism of heterosis utilization in maize improvement. Genome Biology, 2021, 22(1):148.
doi: 10.1186/s13059-021-02370-7 |
| [28] |
Meng L J, Wang B X, Zhao X Q, et al. Association mapping of ferrous,zinc,and aluminum tolerance at the seeding stage in Indica rice using MAGIC populations. Frontiers in Plant Science, 2017, 8:1822.
doi: 10.3389/fpls.2017.01822 |
| [29] | Meng L J, Guo L B, Ponce K, et al. Characterization of three Indica rice multiparent advanced generation inter-cross (magic) populations for quantitative trait loci (QTL) identification. The Plant Genome, 2016, 9(2):1-14. |
| [30] |
Ponce K, Zhang Y, Guo L B, et al. Genome-wide association study of grain size traits in Indica rice multiparent advanced generation intercross (MAGIC) population. Frontiers in Plant Science, 2020, 11:395.
doi: 10.3389/fpls.2020.00395 |
| [31] |
Thépot S, Restoux G, Goldringer I, et al. Effciently tracking selection in a multiparental population:the case of earliness in wheat. Genetics, 2015, 199(2):609-621.
doi: 10.1534/genetics.114.169995 |
| [32] |
Bandillo N, Raghavan C, Muyco P A, et al. Multi-parent advanced generation inter-cross (MAGIC) populations in rice:progress and potential for genetics research and breeding. Rice, 2013, 6(1):1-15.
doi: 10.1186/1939-8433-6-1 |
| [33] | 陈天晓, 朱亚军, 密雪飞, 等. 利用水稻MAGIC群体关联定位白叶枯病抗性QTL和创制抗病新种质. 作物学报, 2016, 42(10):1437-1447. |
| [34] | Raghavan C, Mauleon R, Lacorte V, et al. Approaches in characterizing genetic structure and mapping in a rice multiparental population. G3:Genes, Genomes,Genetics, 2017, 7(6):1721-1730. |
| [35] | Huang B E, Georde A W, Forrest K L, et al. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnology, 2012, 10(7):826-839. |
| [36] |
Rebetzke G J, Verbyla A P, Verbyla K L, et al. Use of a large multiparent wheat mapping population in genomic dissection of coleoptile and seedling growth. Plant Biotechnology Journal, 2014, 12(2):219-230.
doi: 10.1111/pbi.12130 pmid: 24151921 |
| [37] | Mackay I J, Barber T, Bentley A R, et al. An eight parent multiparent advanced generation inter-cross population for winter-sown wheat:creation,properties,and validation. G3:Genes, Genomes,Genetics, 2014, 4(9):1603-1610. |
| [38] |
Malosetti M, Ribaut J M, Eeuwijk F A. The statistical analysis of multi-environment data:modelling genotype-by-environment interaction and its genetic basis. Frontiers in Physiology, 2013, 4:44.
doi: 10.3389/fphys.2013.00044 pmid: 23487515 |
| [39] | 纪耀勇. 高筋全麦粉用途小麦的MAGIC群体法培育及品质检测. 镇江:江苏大学, 2019. |
| [40] |
Thépot S, Restoux G, Goldringer I, et al. Effciently tracking selection in a multiparental population:the case of earliness in wheat. Genetics, 2015, 199(2):609-621.
doi: 10.1534/genetics.114.169995 |
| [41] |
Sannemann W, Huang B E, Mathew B, et al. Multi-parent advanced generation intercross in barley:high-resolution quantitative trait locus mapping for flowering time as a proof of concept. Molecular Breeding, 2015, 35(3):86.
doi: 10.1007/s11032-015-0284-7 |
| [42] | Cockram J, Scuderi A, Barber T, et al. Fine-mapping the wheat Snn1 locus conferring sensitivity to the Parastagonospora nodorum necrotrophic effector SnTox 1 using an eight founder multiparent advanced generation inter-cross population. G3:Genes, Genomes,Genetics, 2015, 5(11):2257-2266. |
| [43] |
Islam M S, Thyssen G N, Jenkins J N, et al. A MAGIC population-based genome-wide association study reveals functional association of GhRBB1_A07 gene with superior fiber quality in cotton. BMC Genomics, 2016, 17(1):903.
doi: 10.1186/s12864-016-3249-2 |
| [44] | Huang C, Shen C, Wen T W, et al. Genome-wide association mapping for agronomic traits in an 8-way Upland cotton MAGIC population by SLAF-seq. Theoretical and Applied Genetics, 2021, 134(8):999-1017. |
| [45] |
Abdelraheem A, Thyssen G N, Fang D D, et al. GWAS reveals consistent QTL for drought and salt tolerance in a MAGIC population of 550 lines derived from intermating of 11 Upland cotton (Gossypium hirsutum) parents. Molecular Genetics and Genomics, 2020, 296(10):119-129.
doi: 10.1007/s00438-020-01733-2 |
| [46] | Ongom P O, Ejeta G. Mating design and genetic structure of a multi-parent advanced generation intercross (MAGIC) population of Sorghum (Sorghum bicolor (L.) Moench). G3:Genes, Genomes, Genetics, 2018, 8(1):331-341. |
| [47] | 刘洪泰. 基于MAGIC群体的烟草黑胫病抗性遗传分析. 北京: 中国农业科学院, 2020. |
| [48] |
Dell'Acqua M, Gatti D M, Pea G, et al. Genetic properties of the MAGIC maize population:a new platform for high definition QTL mapping in Zea mays. Genome Biology, 2015, 16(1):167.
doi: 10.1186/s13059-015-0716-z |
| [49] | Anderson II S L, Mahan A L, Murray S C, et al. Four parent maize (FPM) population:effects of mating designs on linkage disequillibrium and mapping quantitative traits. Plant Genome. 2018, 11(2):1-17. |
| [50] |
Septiani P, Lanubile A, Stagnati L, et al. Unravelling the genetic basis of Fusariom seedling rot resistance in the MAGIC maize population:novel targets for breeding. Scientific Reports, 2019, 9(1):5665.
doi: 10.1038/s41598-019-42248-0 pmid: 30952942 |
| [51] |
Butrón A, Santiago R, Cao A, et al. QTLs for resistance to fusarium ear rot in a multiparent advanced generation intercross (MAGIC) maize population. Plant Disease, 2019, 103(5):897-904.
doi: 10.1094/PDIS-09-18-1669-RE pmid: 30856072 |
| [52] |
Yi Q, Malvar R A, Álvarez-Iglesias L, et al. Dissecting the genetics of cold tolerance in a multiparental maize population. Theoretical and Applied Genetics, 2020, 133(2):503-516.
doi: 10.1007/s00122-019-03482-2 pmid: 31740990 |
| [53] |
Li H, Peng Z, Yang X, et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nature Genetics, 2013, 45(1):43-50.
doi: 10.1038/ng.2484 |
| [54] | Yang N, Lu Y L, Yang X H, et al. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genetics, 2014, 10(9):e1004573. |
| [55] | Giraud H, Bauland C, Falque M, et al. Linkage analysis and association mapping qtl detection models for hybrids between multiparental populations from two heterotic groups:application to biomass production in maize(Zea mays L.). G3:Genes, Genomes,Genetics, 2017, 7(11):3649-3657. |
| [56] | 赵福永, 赵恒, 王晓玲, 等. 甘蓝型油菜MAGIC群体构建及其在遗传育种中的应用潜力. 中国油料作物学报, 2017, 39(2):145-151. |
| [57] |
Mackay I, Powell W. Methods for linkage disequilibrium mapping in crops. Trends in Plant Science, 2007, 12(2):57-63.
doi: 10.1016/j.tplants.2006.12.001 pmid: 17224302 |
| [58] | 郭志军, 赵云雷, 陈伟, 等. 陆地棉SSR标记遗传多样性及其与农艺性状的关联分析. 棉花学报, 2014, 26(5):420-430. |
| [59] | 石治鹏. 棉花MAGIC群体后代株系对缩节胺的敏感性研究. 荆州:长江大学, 2018. |
| [60] | 黄聪. 基于自然群体及MAGIC群体关联分析解析陆地棉重要农艺性状的遗传基础. 武汉:华中农业大学, 2018. |
| [61] |
Liang Y M, Liu H J, Yan J B, et al. Natural variation in crops:realized understanding,continuing promise. Annual Review of Plant Biology, 2021, 72:357-385.
doi: 10.1146/annurev-arplant-080720-090632 |
| [62] | Yu J, Wang J, Lin W, et al. The genomes of Oryza sativa:a history of duplications. PLoS Biology, 2005, 3(2):266-281. |
| [63] |
Giraud H, Lehermeier C, Bauer E, et al. Linkage disequilibrium with linkage analysis of multiline crosses reveals different multiallelic QTL for hybrid performance in the flint and dent heterotic groups for maize. Genetics, 2014, 198(4):1717-1734.
doi: 10.1534/genetics.114.169367 |
| [64] |
Consortium C C. The genome architecture of the colaborative cross mouse genetic reference population. Genetics, 2012, 190(2):389-402.
doi: 10.1534/genetics.111.132639 |
| [65] |
Verbyla A P, George A W, Cavanagh C R, et al. Whole-genome QTL analysis for MAGIC. Theoretical and Applied Genetics, 2014, 127(8):1753-1770.
doi: 10.1007/s00122-014-2337-4 pmid: 24927820 |
| [66] |
Zhao Y L, Wang H M, Chen W, et al. Genetic diversity and population structure of elite cotton(Gossypium hirsutum L.) germplasm revealed by SSR markers. Plant Systematics and Evolution, 2015, 301(1):327-336.
doi: 10.1007/s00606-014-1075-z |
| [67] |
Scutari M, Howell P, Balding D J, et al. Multiple quantitative trait analysis using bayesian networks. Genetics, 2014, 198(1):129-137.
doi: 10.1534/genetics.114.165704 pmid: 25236454 |
| [68] |
Corbett-Detig R B, Zhou J, Clark A G, et al. Genetic incompatibilities are widespresd within species. Nature, 2013, 504(7478):135-137.
doi: 10.1038/nature12678 |
| [69] |
Huang B E, Verbyla K L, Verbyla A P, et al. MAGIC populations in crops:current status and future prospects. Theoretical and Applied Genetics, 2015, 128(6):999-1017.
doi: 10.1007/s00122-015-2506-0 pmid: 25855139 |
| [70] |
Hemani G, Theocharidis A, Wei W H, et al. EpiGPU:exhaustive pairwise epistasis scans parallelized on consumer level graphics cards. Bioinformatics, 2011, 27(11):1462-1465.
doi: 10.1093/bioinformatics/btr172 |
| [71] | Gyenesei A, Semple CM, Haley C S, et al. High throughput analysis of epistasis in genome-wide association studies with BiForce. Bioinformatics, 2013, 29(20):1957-1964. |
| [1] | Wu Jianzhong,Li Suiyan,Lin Hong,Ma Yanhua,Pan Liyan,Li Donglin,Sun Dequan. Genetic Variation and Principal Component Analysis of Silage Maize Quality Traits [J]. Crops, 2019, 35(3): 42-48. |
| [2] | Huan Wang,Bing Liu,Ziyu Bai,Junli Shi,Jinlin Ye. Effects of Laser Irradiation and Hybridization on the DNA Methylation Alteration in Sorghum (Sorghum bicolor L.) [J]. Crops, 2017, 33(1): 32-37. |
|
||