Crops ›› 2023, Vol. 39 ›› Issue (5): 71-80.doi: 10.16035/j.issn.1001-7283.2023.05.011
Previous Articles Next Articles
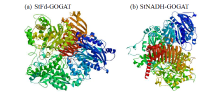
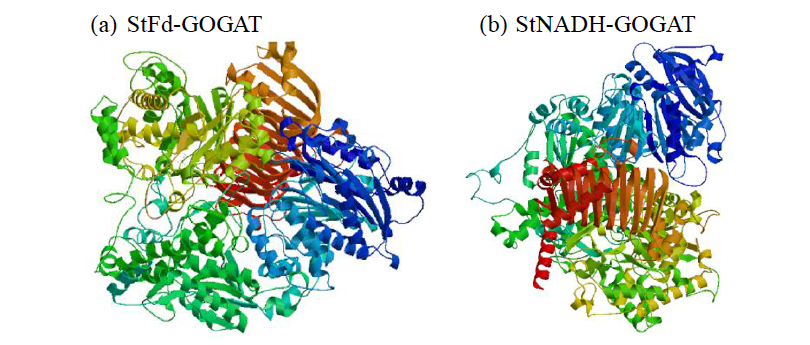
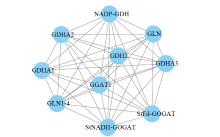
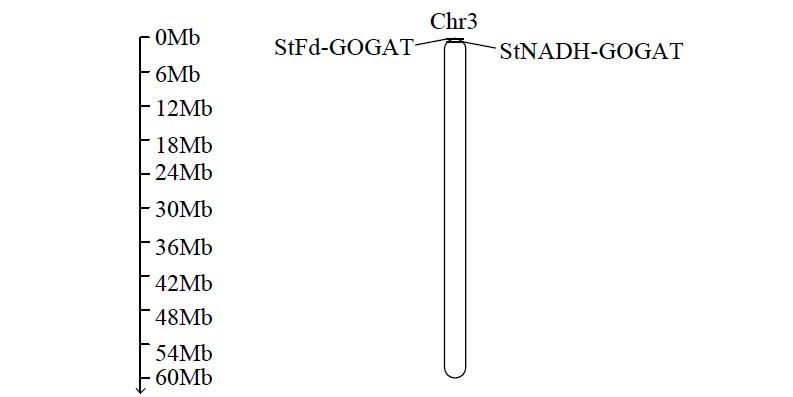
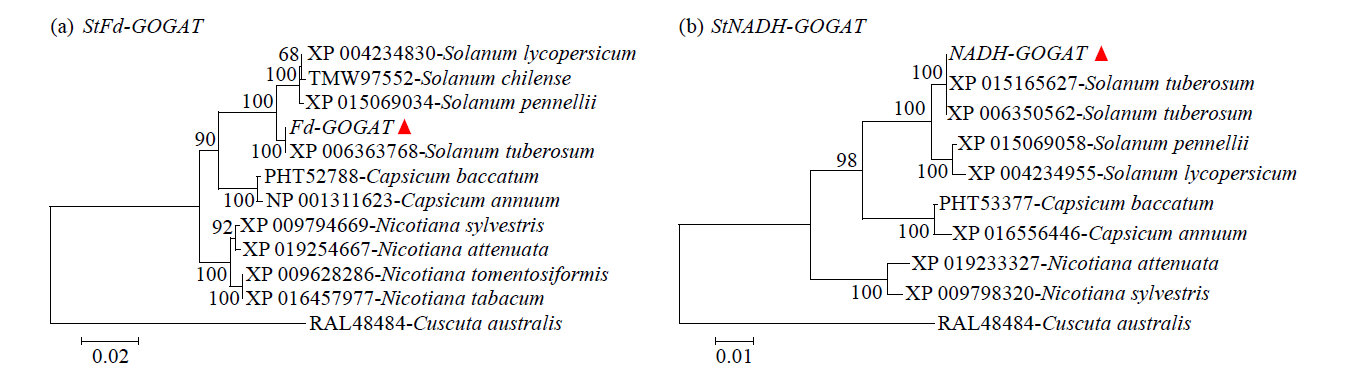
Cloning and Analysis of the Structure and Function of the Key Enzyme Genes StGOGATs of Potato Nitrogen Assimilation
Han Zhijun1( ), Zhao Yanfei1, Lu Yue1, Li Shuang2, Zhang Jiayue1, Han Yuzhu1(
), Zhao Yanfei1, Lu Yue1, Li Shuang2, Zhang Jiayue1, Han Yuzhu1( ), Zhang Jingying1(
), Zhang Jingying1( )
)
- 1College of Horticulture, Jilin Agricultural University, Changchun 130118, Jilin, China
2Teaching and Research Base Management Office, Jilin Agricultural University, Changchun 130118, Jilin, China
| [1] | 郝凯, 贾立国, 秦永林, 等. 氮素对马铃薯源-库关系影响研究进展. 作物杂志, 2020(3):22-26. |
| [2] | 张静, 蒙美莲, 王颖慧, 等. 氮磷钾施用量对马铃薯产量及品质的影响. 作物杂志, 2012(4):124-127. |
| [3] |
Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, et al. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Annals of Botany, 2010, 105(7):1141-1157.
doi: 10.1093/aob/mcq028 pmid: 20299346 |
| [4] |
McAllister C H, Beatty P H, Good A G. Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnology Journal, 2012, 10(9):1011-1025.
doi: 10.1111/j.1467-7652.2012.00700.x pmid: 22607381 |
| [5] | 刘秀杰, 宫占元. 植物氮素吸收利用研究进展. 现代化农业, 2012(8):20-21. |
| [6] | 马雪峰, 高旻, 程治军. 植物氮素吸收与利用的分子机制研究进展. 作物杂志, 2013(4):32-38. |
| [7] | 张婷婷, 孟丽丽, 陈有君, 等. 不同马铃薯品种的氮效率差异研究. 中国土壤与肥料, 2021(1):63-69. |
| [8] | Fontaine J X, Tercélaforgue T, Armengaud P, et al. Characterization of a NADH-Dependent glutamate dehydrogenase mutant of demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Bulletin of the American Physical Society, 2014. |
| [9] | 张华珍, 徐恒玉. 植物氮素同化过程中相关酶的研究进展. 北方园艺, 2011(20):180-183. |
| [10] | 牛超, 刘关君, 曲春浦, 等. 谷氨酸合成酶基因及其在植物氮代谢中的调节作用综述. 江苏农业科学, 2018, 46(9):10-16. |
| [11] |
Konishi N, Ishiyama K, Matsuoka K, et al. NADH-dependent glutamate synthase plays a crucial role in assimilating ammonium in the Arabidopsis root. Physiologia Plantarum, 2014, 152(1):138-151.
doi: 10.1111/ppl.12177 |
| [12] | 朱静, 岳思宁, 陈琛, 等. 谷氨酸合酶在灵芝中生物学功能的研究. 南京农业大学学报, 2019, 42(6):1073-1079. |
| [13] | 陈丽华, 刘丽君, 刘页丽, 等. 不同基因型大豆Fd-GOGAT基因cDNA序列的克隆与分析. 大豆科学, 2011, 30(3):374-378. |
| [14] |
陈阳, 孙华山, 王玉书, 等. 草地早熟禾NADH-GOGAT基因的克隆及表达分析. 草地学报, 2019, 27(2):459-465.
doi: 10.11733/j.issn.1007-0435.2019.02.026 |
| [15] | 张云, 赵艳菲, 王雅平, 等. 延薯4号马铃薯对氮素的生理生化响应及转录组分析. 广东农业科学, 2021, 48(2):56-66. |
| [16] | 赵艳菲, 张嘉越, 韩玉珠. 马铃薯氮代谢途径中Fd-GOGAT基因的克隆及生物信息学分析//中国作物学会马铃薯专业委员会,马铃薯产业与美丽乡村(2020). 哈尔滨: 黑龙江科学技术出版社, 2020. |
| [17] |
Kishorekumar R, Bulle M, Wany A, et al. An overview of important enzymes involved in nitrogen assimilation of plants. Methods in Molecular Biology, 2020, 2057:1-13.
doi: 10.1007/978-1-4939-9790-9_1 pmid: 31595465 |
| [18] |
Bernard S M, Habash D Z. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytologist, 2009, 182(3):608-620.
doi: 10.1111/j.1469-8137.2009.02823.x pmid: 19422547 |
| [19] | 黄成能. N胁迫对柑橘营养生长及GS/GOGAT循环酶基因表达影响的研究. 长沙:湖南农业大学, 2014. |
| [20] |
Clough S J, Bent A F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal, 1998, 16(6):735-743.
doi: 10.1046/j.1365-313x.1998.00343.x pmid: 10069079 |
| [21] |
Feraud M, Masclaux-Daubresse C, Ferrario-Méry S, et al. Expression of a ferredoxin-dependent glutamate synthase gene in mesophyll and vascular cells and functions of the enzyme in ammonium assimilation in Nicotiana tabacum (L.). Planta, 2005, 222(4):667-677.
doi: 10.1007/s00425-005-0013-2 |
| [22] |
Lea P J, Miflin B J. Alternative route for nitrogen assimilation in higher plants. Nature, 1974, 251(5476):614-616.
doi: 10.1038/251614a0 |
| [23] |
Wallsgrove R M, Lea P J, Miflin B J. Distribution of the enzymes of nitrogen assimilation within the pea leaf cell. Plant Physiology, 1979, 63(2):232-236.
doi: 10.1104/pp.63.2.232 pmid: 16660703 |
| [24] |
Becker T W, Carrayol E, Hirel B. Glutamine synthetase and glutamate dehydrogenase isoforms in maize leaves: localization,relative proportion and their role in ammonium assimilation or nitrogen transport. Planta, 2000, 211(6):800-806.
pmid: 11144264 |
| [25] |
Loulakakis K A, Primikirios N I, Nikolantonakis M A, et al. Immunocharacterization of Vitis vinifera L. ferredoxin-dependent glutamate synthase,and its spatial and temporal changes during leaf development. Planta, 2002, 215(4):630-638.
doi: 10.1007/s00425-002-0785-6 |
| [26] |
Zeng D D, Qin R, Li M, et al. The ferredoxin-dependent glutamate synthase (OsFd-GOGAT) participates in leaf senescence and the nitrogen remobilization in rice. Molecular Genetics and Genomics, 2017, 292(2):385-395.
doi: 10.1007/s00438-016-1275-z |
| [27] | 张翼飞. 施氮对甜菜氮素同化与碳代谢的调控机制研究. 哈尔滨:东北农业大学, 2013. |
| [28] |
侯昕, 徐新翔, 贾志航, 等. 供氮水平对苹果砧木‘M9T337’幼苗生长和GS、GOGAT、AS基因表达的影响. 园艺学报, 2019, 46(11):2239-2248.
doi: 10.16420/j.issn.0513-353x.2018-1010 |
| [29] |
Yamaya T, Obara M, Nakajima H, et al. Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. Journal of Experimental Botany, 2002, 53:917-925.
doi: 10.1093/jexbot/53.370.917 pmid: 11912234 |
| [1] | Cao Liru, Wang Guorui, Zhang Xin, Wei Liangming, Wei Xin, Zhang Qianjin, Deng Yazhou, Wang Zhenhua, Lu Xiaomin. Genome-Wide Identification and Analysis of HSP90 Gene Family in Maize [J]. Crops, 2021, 37(5): 28-34. |
| [2] | Wang Li, Wang Zuoping, Zhang Zhongbao, Bai Ling, Wu Zhongyi. Screening of Strongly Expressed Promoters in Immature Maize Kernels [J]. Crops, 2020, 36(4): 114-120. |
| [3] | Li Yan,Qiang Yang,Yupeng Shao,Dandan Li,Zhikun Wang,Wenbin Li. Cloning and Sequence Analysis of GmWRI1a Gene Promoter in Soybean [J]. Crops, 2017, 33(2): 51-58. |
| [4] | Liu Jianwei, Chen Xiaofeng, Liu Guangfu, Guo Zongduan, Li Xinzhu, Hu Zhaoping, Zhang liang. Cloning and Expression Analysis of CYP78A5,a Tissue-specific Promoter in Soybean [J]. Crops, 2014, 30(1): 54-58. |
|
||