Crops ›› 2024, Vol. 40 ›› Issue (3): 13-22.doi: 10.16035/j.issn.1001-7283.2024.03.003
Previous Articles Next Articles
Transcriptome Analysis of Different Foxtail Millet (Setaria italica L.) Varieties Treated with Imazapic Herbicide
Song Hui1( ), Wang Tao2, Xing Lu1, Liu Junfang1, Zhang Yang1, Liu Jinrong1, Chen Hongqi1(
), Wang Tao2, Xing Lu1, Liu Junfang1, Zhang Yang1, Liu Jinrong1, Chen Hongqi1( ), Feng Baili3
), Feng Baili3
- 1Anyang Academy of Agricultural Sciences, Anyang 455000, Henan, China
2College of Biology and Food Engineering, Anyang Institute of Technology, Anyang 455000, Henan, China
3College of Agriculture, Northwest Agriculture and Forestry University / State Key Laboratory of Crop Stress Biology in Arid Areas, Yangling 712100, Shaanxi, China
| [1] | 刁现民. 基础研究提升传统作物谷子和黍稷的科研创新水平. 中国农业科学, 2016, 49(17):3260-3262. |
| [2] |
Muthamilarasan M, Prasad M. Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theoretical and Applied Genetics, 2015, 128(1):1-14.
doi: 10.1007/s00122-014-2399-3 pmid: 25239219 |
| [3] |
刘宝玲, 张莉, 孙岩, 等. 谷子bZIP转录因子的全基因组鉴定及其在干旱和盐胁迫下的表达分析. 植物学报, 2016, 51(4):473-487.
doi: 10.11983/CBB15148 |
| [4] | 周汉章, 王新玉, 薄奎勇, 等. 夏谷田阔叶杂草密度与谷子产量损失关系的研究. 作物杂志, 2013(1):108-112. |
| [5] | Dayan F E, Zaccaro M L D M. Chlorophyll fluorescence as a marker for herbicide mechanisms of action. Pesticide Biochemistry and Physiology, 2012, 102:189-197. |
| [6] | Jimenez F, Fernandez P, Rojano-Delgado A M, et al. Resistance to imazamox in Clearfield soft wheat (Triticum aestivum L.). Crop Protection, 2015, 78:15-19. |
| [7] |
Reddy K N, Bellaloui N, Zablotowicz R M. Glyphosate effect on shikimate, nitrate reductase activity, yield, and seed composition in corn. Journal of Agricultural and Food Chemistry, 2010, 58(6):3646-3650.
doi: 10.1021/jf904121y pmid: 20180575 |
| [8] | Green J M. Review of glyphosate and als-inhibiting herbicide crop resistance and resistant weed management. Weed Technology, 2007, 21(2):547-558. |
| [9] |
Green J M, Owen M D K. Herbicide-resistant crops: Utilities and limitations for herbicide-resistant weed management. Journal of Agricultural and Food Chemistry, 2011, 59(11):5819-5829.
doi: 10.1021/jf101286h pmid: 20586458 |
| [10] |
Green J M. The benefits of herbicide-resistant crops. Pest Management Science, 2012, 68(10):1323-1331.
doi: 10.1002/ps.3374 pmid: 22865693 |
| [11] | Breccia G, Gil M, Vega T, et al. Contribution of non-target-site resistance in imidazolinones-resistant Imisun sunflower. Bragantia, 2017, 76(4):536-542. |
| [12] | Edwards R, Cole D J. Glutathione transferases in wheat (Triticum) species with activity toward fenoxaprop-ethyl and other herbicides. Pesticide Biochemistry and Physiology, 1996, 54:96-104. |
| [13] | Hatton P J, Dixon D, Cole D J, et al. Glutathione transferase activity and herbicide selectivity in maize and associated weed species. Pest Management Science, 1996, 46:267-275. |
| [14] | Cummins I, Wortley D J, Sabbadin F, et al. Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(15):5812-5817. |
| [15] | Kebeish R, Azab E, Peterhaensel C, et al. Engineering the meta- bolism of the phenylurea herbicide chlortoluron in genetically modified Arabidopsis thaliana plants expressing the mammalian cytochrome P450 enzyme CYP1A2. Environmental Science and Pollution Research, 2014, 21:8224-8232. |
| [16] |
宋慧, 王涛, 田礼新, 等. 谷子抗咪唑啉酮的遗传应用和基因初定位. 中国农学通报, 2021, 37(28):1-8.
doi: 10.11924/j.issn.1000-6850.casb2020-0749 |
| [17] | Martin M. Cutadapt removes adapter sequences from high- throughput sequencing reads. Embnet Journal, 2011, 17(1):10-12. |
| [18] |
Kim D, Langmead B, Salzberg S L. HISAT: a fast spliced aligner with low memory requirements. Nature Methods, 2015, 12(4):357-360.
doi: 10.1038/nmeth.3317 pmid: 25751142 |
| [19] |
Kim D, Pertea G, Trapnell C, Pimentel H, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 2013, 14(4):36.
doi: 10.1186/gb-2013-14-4-r36 pmid: 23618408 |
| [20] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods in Molecular Biology, 2011, 25:402-408. |
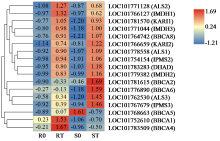
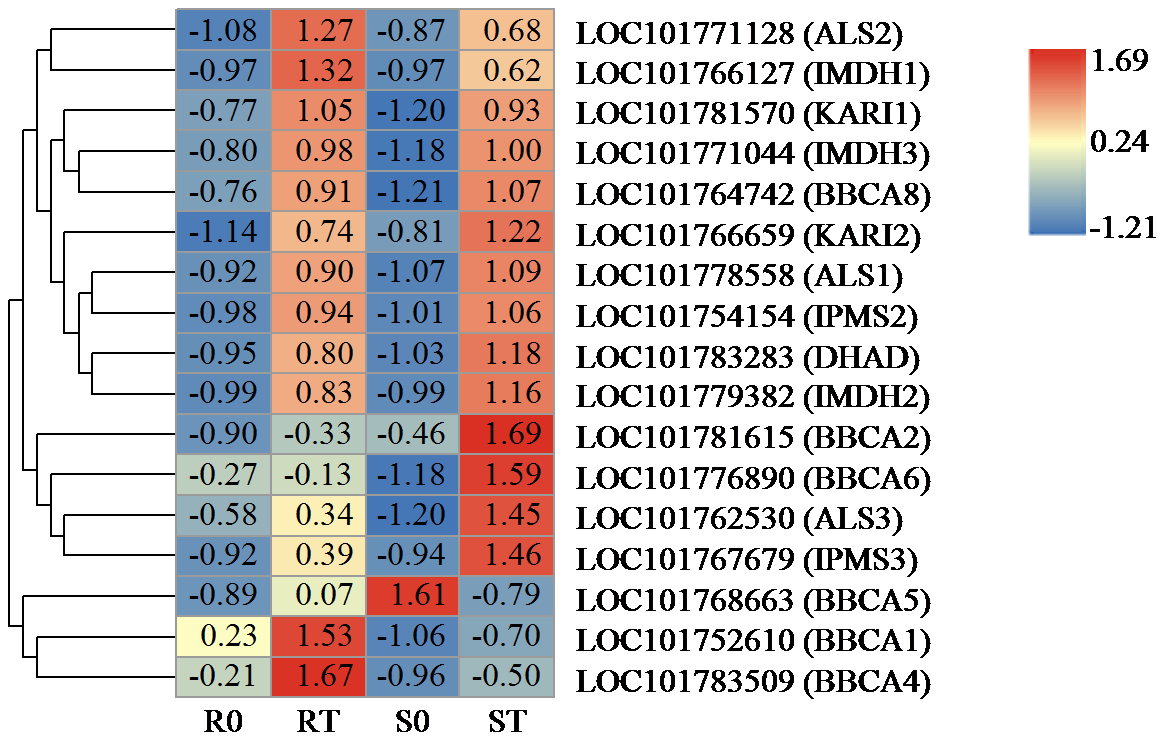
| [21] | Chen H, Saksa K, Zhao F, et al. Genetic analysis of pathway regulation for enhancing branched-chain amino acid biosynthesis in plants. Plant Journal, 2010, 63(4):573-583. |
| [22] | Xing A, Last R L. A regulatory hierarchy of the arabidopsis branched-chain amino acid metabolic network. Plant Cell, 2017, 29(6):1480-1499. |
| [23] |
Ochogavía A C, Gil M, Picardi L, et al. Precision phenotyping of imidazolinones-induced chlorosis in sunflower. Breeding Science, 2014, 64:416-421.
doi: 10.1270/jsbbs.64.416 pmid: 25914598 |
| [24] |
Balabanova D A, Paunov M, Goltsev V, et al. Photosynthetic performance of the imidazolinones resistant sunflower exposed to single and combined treatment by the herbicide imazamox and an amino acid extract. Frontiers in Plant Science, 2016, 7:1559.
pmid: 27826304 |
| [25] |
Jez J M, Noel J P. A kaleidoscope of carotenoids. Nature Biotechnology, 2000, 18(8):825-826.
pmid: 10932147 |
| [26] | Kim B H, Kim S Y, Nam K H. Genes encoding plant-specific class III peroxidases are responsible for increased cold tolerance of the brassinosteroid-insensitive 1mutant. Molecules & Cells, 2012, 34(6):539-548. |
| [27] | Hedtke B, Alawady A, Albacete A, et al. Deficiency in riboflavin biosynthesis affects tetrapyrrole biosynthesis in etiolated Arabidopsis tissue. Plant Molecular Biology, 2012, 78(1/2):77-93. |
| [28] | Qin Y M, Hu CY, Pang Y, et al. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation. Plant Cell, 2007, 19(11):3692-3704. |
| [29] | Denic V, Weissman J S. Molecular caliper mechanism for determining very-long chain fatty acid length. Cell, 2007, 130:663-677. |
| [1] | Du Jie, Su Fuli, Xia Qing, Zhi Hui, Wang Wenxia. Physiological and Biochemical Responses of Foxtail Millet to Low Temperature Stress at Seedling Stage [J]. Crops, 2024, 40(3): 180-185. |
| [2] | Liu Ying, Yin Zequn, Wu Baichen, Xu Mingli, Liu Chang, Shi Huishu, Pang Bo, Miao Xingfen. Effects of Compound Saline-Alkali Stress on Germination Period of Different Foxtail Millet Varieties and Screening of Saline-Alkali Tolerance Varieties [J]. Crops, 2024, 40(3): 207-215. |
| [3] | Qing Chen, Liu Zhengxue, Li Yanjie. Effects of Compound Microbial Fertilizer on Drought Resistance of Maize Seedlings under Drought Stress by Transcriptome Analysis [J]. Crops, 2024, 40(3): 32-39. |
| [4] | Dou Weize, Jiang Wen, Feng Yichuan, Jin Zhuo, Quan Xueli, Wu Songquan. Transcriptome Analysis Reveals Mechanisms of Calycosin- 7-O-β-D-Glucoside Accumulation in Astragalus membranaceus Adventitious Roots through Hydrogen Peroxide [J]. Crops, 2024, 40(1): 48-56. |
| [5] | Dong Haosheng, Wang Qi, Yan Peng, Xu Yanli, Zhang Wei, Lu Lin, Dong Zhiqiang. Effects of ECK on the Lodging Resistance and Yield of Foxtail Millet Stem [J]. Crops, 2023, 39(6): 181-189. |
| [6] | Zhao Lijie, Zhao Haiyan, Han Genlan, Wang Jiang, Nie Mengʼen, Du Huiling, Yuan Xiangyang, Dong Shuqi. Effects of Nitrogen Fertilizer Combined with Organic Fertilizer on Quality of Millet [J]. Crops, 2023, 39(6): 224-232. |
| [7] | Shen Tianyu, Wang Yuan, Dong Erwei, Wang Jinsong, Liu Qiuxia, Jiao Xiaoyan. Effects of Nitrogen and Delayed Harvest on Foxtail Millet Yield and Grain Quality [J]. Crops, 2023, 39(6): 233-242. |
| [8] | Zhao Haiyan, Zhao Lijie, Han Genlan, Wang Jiang, Wang Zijian, Nie Meng’en, Du Huiling, Yuan Xiangyang, Dong Shuqi. Effects of Nitrogen and Zinc Application on Root Morphology and Zinc Content in Foxtail Millet [J]. Crops, 2023, 39(4): 152-158. |
| [9] | Liu Songtao, Tian Zaimin, Liu Zigang, Gao Zhijia, Zhang Jing, He Donggang, Huang Zhihong, Lan Xin. Transcriptomic Analysis to Reveal Lodging Resistance Genes and Metabolism Pathways in Maize (Zea mays L.) [J]. Crops, 2023, 39(4): 31-37. |
| [10] | Zhao Yun, Feng Guojun, Hu Xiangwei, Wumaierjiang·Kuerban , Li Pengbing, Li Cuimei, Akebota·Muheyati . Preliminary Report on Selection of Herbicide-Resistant Foxtail Millet Varieties Suitable for Planting in Kashgar, Xinjiang [J]. Crops, 2023, 39(3): 126-133. |
| [11] | He Shuiling, Zhao Xia, Wu Mingqi, Wang Dongsheng. Effects of Exogenous Nitric Oxide and Hydrogen Sulfide on the Germination of Foxtail Millet Seeds [J]. Crops, 2023, 39(2): 138-144. |
| [12] | Ma Jiyu, Wang Shuang, Li Yun, Guo Zhenqing, Wang Jian, Lin Xiaohu, Han Yucui. Effects of Planting Density on Agronomic Characteristics and Yield of Foxtail Millet [J]. Crops, 2023, 39(2): 222-228. |
| [13] | Wang Huimin, Li Minghao, Li Yun, Li Han, Wang Shuang, Lin Xiaohu, Han Yucui. Identification and Evaluation of Salt-Alkali Tolerance of Foxtail Millet Cultivars (Lines) at Germination Stage [J]. Crops, 2023, 39(2): 57-66. |
| [14] | Bian Shuhui, Xing Guofang, Liang Xin, Zhang Shuwei, Wang Jianing, Ye Haoyu. Effects of Different Forms Selenium and Dosage on Foxtail Millet Growth and Physiology at Seedling Stage [J]. Crops, 2023, 39(1): 152-157. |
| [15] | Wang Qi, Xu Yanli, Yan Peng, Dong Haosheng, Zhang Wei, Lu Lin, Dong Zhiqiang. Effects of Polyaspartic Acid-Chitosan on Agronomic Traits, Yield and Nitrogen Use of Spring Foxtail Millet [J]. Crops, 2023, 39(1): 58-67. |
|
||