作物杂志,2016, 第4期: 62–67 doi: 10.16035/j.issn.1001-7283.2016.04.010
亚麻糖基转移酶基因LuUGT72E1的克隆与表达分析
袁红梅1,2,郭文栋3,赵丽娟4,于莹2,吴建忠2,程莉莉2,赵东升2,康庆华2,黄文功2,姚玉波2,宋喜霞2,姜卫东2,刘岩2,马廷芬2,吴广文2,关凤芝2
- 1 黑龙江省农业科学院博士后工作站,150086,黑龙江哈尔滨
2 黑龙江省农业科学院经济作物研究所,150086,黑龙江哈尔滨
3 黑龙江省科学院自然与生态研究所,150040,黑龙江哈尔滨
4 黑龙江省农业科学院作物育种所,150086,黑龙江哈尔滨
Cloning and Expression Analysis of the Glycosyltransferase Gene LuUGT72E1 in Flax
Yuan Hongmei1,2,Guo Wendong3,Zhao Lijuan4,Yu Ying2,Wu Jianzhong2,Cheng Lili2,Zhao Dongsheng2,Kang Qinghua2,Huang Wengong2,Yao Yubo2,Song Xixia2,Jiang Weidong2,Liu Yan2,Ma Tingfen2,Wu Guangwen2,Guan Fengzhi2
- 1 Heilongjiang Academy of Agricultural Sciences Postdoctoral Programme,Harbin 150086,Heilongjiang,China
2 Industrial Crops Institute,Heilongjiang Academy of Agricultural Sciences,Harbin 150086,Heilongjiang,China
3 Nature and Ecology Institute,Heilongjiang Academy of Sciences,Harbin 150040,Heilongjiang,China
4 Crop Breeding Institute,Heilongjiang Academy of Agricultural Sciences,Harbin 150086,Heilongjiang,China
摘要:
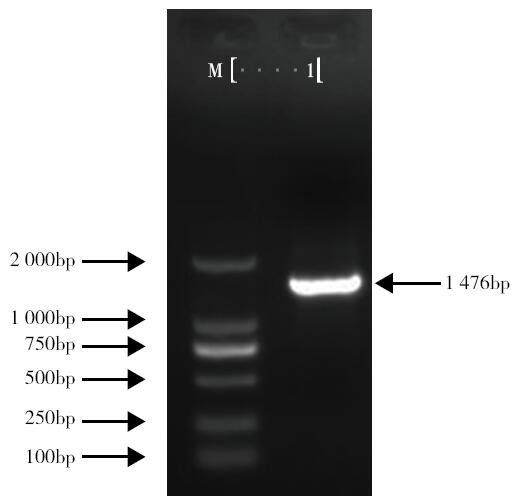
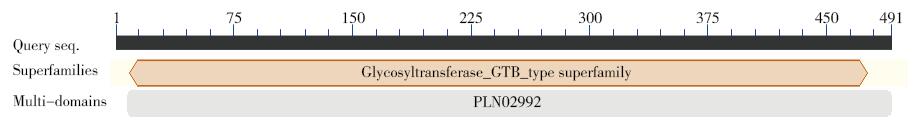
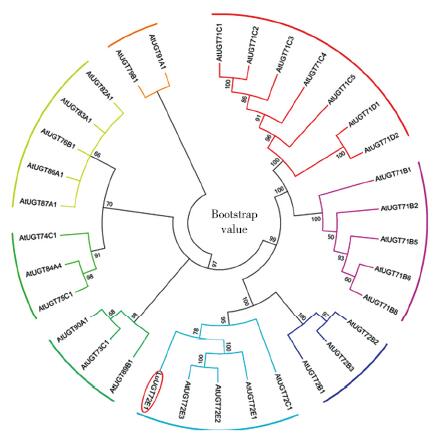
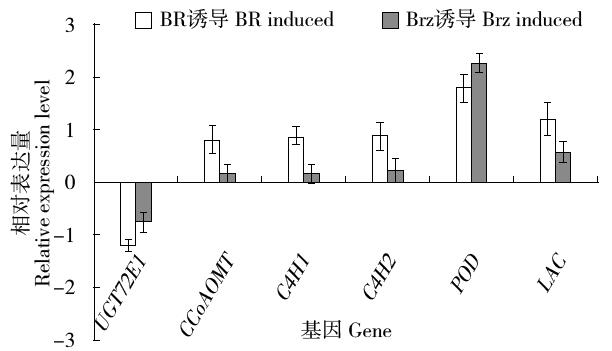
以亚麻茎部组织总RNA为模版,通过反转录PCR获得糖基转移酶LuUGT72E1基因的全长开放读码框序列。序列分析结果表明:开放读码框全长1 476bp,共编码491个氨基酸。通过多氨基酸比对发现,该蛋白与亚麻UGT72N2蛋白亲缘关系最近,相似度达99%,与毛果杨、巨桉、木薯、雷蒙德氏棉及拟南芥UGT72E1蛋白的氨基酸序列相似性在58%~51%。分子进化分析表明,该蛋白与拟南芥UGT72E1~3聚为一类,推测该蛋白参与木质素单体的糖基化修饰过程。基因表达谱结果表明,该基因被BR、Brz诱导下调表达。本研究为亚麻的纤维品质改良提供了候选基因。
| [1] | Franke R, Hemm M R, Denault J W , et al. Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Planta, 2002,30(1):47-59. |
| [2] |
Novaes E, Kirst M, Chiang V , et al. Lignin and biomass:a negative correlation for wood formation and lignin content in trees. Plant Physiology, 2010,154(2):555-561.
doi: 10.1104/pp.110.161281 |
| [3] | 魏建华, 宋艳茹 . 木质素生物合成途径及调控的研究进展. 植物学报, 2001,43(8) : 771-779. |
| [4] |
陈晓光, 史春余, 尹燕枰 , 等. 小麦茎秆木质素代谢及其与抗倒性的关系. 作物学报, 2011,37(9):1616-1622.
doi: 10.3724/SP.J.1006.2011.01616 |
| [5] | Jones L, Ennos A R, Turner S R . Cloning and characterization of irregular xylem4 (irx4):A severely lignin-deficient mutant of Arabidopsis. Planta, 2001,26(2):205-216. |
| [6] |
Ruben V, Véronique S, Bartel V , et al. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell, 2012,24(9):3506-3529.
doi: 10.1105/tpc.112.102574 |
| [7] |
Marie B, Bernard M, Marc V M , et al. Biosynthesis and genetic enginering of lignin. Critical Reviews in Plant Sciences, 1998,17(2):125-197.
doi: 10.1080/07352689891304203 |
| [8] |
Barry H . Lignin synthesis:the generation of hydrogen peroxide and superoxide by horseradish peroxidase and its stimulation by manganese(II) and phenols. Planta, 1978,140(1):81-88.
doi: 10.1007/BF00389384 |
| [9] | Du X, Gellerstedt G, Li J . Universal fractionation of lignin-carbohydrate complexes(LCCs) from lignocellulosic biomass:an example using spruce wood. Planta, 2013,74(2):328-338. |
| [10] |
Xu L, Nicholas D B, Jing K W , et al. The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. The Plant Cell, 2010,22(5):1620-1632.
doi: 10.1105/tpc.110.074161 |
| [11] | Du X Y, Marta P B, Carmen F , et al. Analysis of lignin-carbohydrate and lignin-lignin linkages after hydrolase treatment of xylan-lignin,glucomannan-lignin and glucan-lignin complexes from spruce wood. Planta, 2014,4(15):323-358. |
| [12] | Hao Z, Mohnen D . A review of xylan and lignin biosynthesis:Foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes.Critical Reviews Biochemistry Molecular Biology, 2014( doi:10.3109/10409238.2014.889651). |
| [13] | 王艳文 .杨树糖基转移酶与木质素合成关系的转基因研究. 济南:山东大学, 2012. |
| [14] | Lanot A, Hodge D, Jackson R G , et al. The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis. Planta, 2006,48(2):286-295. |
| [15] |
Lanot A, Hodge D, Lim E K , et al. Redirection of flux through the phenylpropanoid pathway by increased glucosylation of soluble intermediates. Planta, 2008,228(4):609-616.
doi: 10.1007/s00425-008-0763-8 |
| [16] |
Baucher M, Bernard-Vailhé M A,Chabbert B, et al. Down-regulation of cinnanyl alcohol dehydrogenase in transgenic alfalfa(Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Molecular Biology, 1999,39(3):437-447.
doi: 10.1023/A:1006182925584 |
| [17] |
Li X C, Richard F H . Synthesis and rearrangement reactions of ester-linked lignin-carbohydrate model compounds. Journal of Agricultural and Food Chemistry, 1995,43(8):2098-2103.
doi: 10.1021/jf00056a026 |
| [18] |
Paquette S, Møller B L, Bak S . On the origin of family 1 plant glycosyltransferases. Phytochemistry, 2003,62(3):399-413.
doi: 10.1016/S0031-9422(02)00558-7 |
| [19] |
Lim E K, Bowles D J . A class of plant glycosyltransferases involved in cellular homeostasis. The EMBO Journal, 2004,23(15):2915-2922.
doi: 10.1038/sj.emboj.7600295 |
| [20] |
Bowles D, Isayenkova J, Lim E K , et al. Glycosyltransferases:Managers of small molecules. Current Opinion in Plant Biology, 2005,8(3):254-263.
doi: 10.1016/j.pbi.2005.03.007 |
| [21] |
Gachon C M, Langlois-Meurinne M, Saindrenan P . Plant secondary metabolism glycosyltransferases:The emerging functional analysis. Trends in Plant Science, 2005,10(11):542-549.
doi: 10.1016/j.tplants.2005.09.007 |
| [22] | Lim E K . Plant glycosyltransferases:Their potential as novel biocatalysts. Chemistry:A European Journal, 2005,11(19):5487-5494. |
| [23] |
Li Y, Baldauf S, Lim E K , et al. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. The Journal of Biological Chemistry, 2001,276(6):4338-4343.
doi: 10.1074/jbc.M007447200 |
| [24] | Ross J, Li Y, Lim E , et al. Higher plant glycosyltransferases. Genome Biology, 2001,2(2):31-36. |
| [1] | 罗海斌, 蒋胜理, 黄诚梅, 曹辉庆, 邓智年, 吴凯朝, 徐林, 陆珍, 魏源文. 甘蔗ScHAK10基因克隆及表达分析[J]. 作物杂志, 2018, (4): 53–61 |
| [2] | 岳德成,史广亮,韩菊红,姜延军,柳建伟,李青梅. 全膜双垄沟播玉米田覆盖化学除草地膜对后茬亚麻生长发育的影响[J]. 作物杂志, 2016, (6): 148–153 |
| [3] | 王树彦,韩冰,周四敏,徐军. 油用亚麻可溶性糖、脂肪含量与硬脂酰-酰基载体蛋白脱氢酶基因表达相关性分析[J]. 作物杂志, 2016, (4): 56–61 |
| [4] | 韩赞平,陈彦惠,郭书磊,祖小峰,王顺喜,赵西拥. 玉米抗逆基因ZmqLTG3-1的克隆及功能分析[J]. 作物杂志, 2016, (4): 47–55 |
| [5] | 李钰,郑文寅,冯春,王荣富,李娟. 非生物逆境胁迫下普通小麦烟农19幼苗FeSOD基因表达分析[J]. 作物杂志, 2016, (4): 75–79 |
| [6] | 姚玉波,吴广文,黄文功,康庆华,姜卫东,路颖,张树权. 不同基因型亚麻钾利用效率差异分析[J]. 作物杂志, 2016, (4): 80–85 |
| [7] | 曹秀霞,钱爱萍,张炜,杨崇庆. 锌肥不同用量对旱地油用亚麻生长及种子产量的影响[J]. 作物杂志, 2016, (3): 167–170 |
| [8] | 郭媛, 邱财生, 龙松华, 等. 种子萌发期亚麻种质资源耐镉性的鉴定评价[J]. 作物杂志, 2015, (6): 39–43 |
| [9] | 郭媛, 邱财生, 龙松华, 等. 福胁迫对不同地区亚麻主栽品种种子萌发的影响[J]. 作物杂志, 2015, (4): 146–151 |
| [10] | 吴鹏, 郭茜茜, 武涛, 等. 黄瓜ABC转运蛋白基因(abca19)的克隆及其对农药霜霉威胁迫的响应[J]. 作物杂志, 2015, (3): 45–51 |
| [11] | 李丹丹, 韩冰, 王树彦, 等. 亚麻子中α-亚麻酸及参与其形成的不饱和脂肪酸的研究进展[J]. 作物杂志, 2015, (2): 18–22 |
| [12] | 吴文荣, 牛瑞明, 苑莹, 等. 外源NO对模拟干旱胁迫下亚麻种子发芽及幼苗生长的影响[J]. 作物杂志, 2015, (1): 143–147 |
| [13] | 吴建忠. 亚麻纤维素合酶关键基因(LusiCesAl)的克隆[J]. 作物杂志, 2014, (6): 36–39 |
| [14] | 陈芳, 党占海, 张建平, 等. 不同基因型亚麻下胚轴不定芽诱导的研究[J]. 作物杂志, 2014, (3): 39–43 |
| [15] | 孟繁君, 陈明, 徐长营, 等. 玉米C2H2型锌指蛋白基因ZFP225的鉴定、生物信息学分析与克隆[J]. 作物杂志, 2014, (1): 49–53 |
|
||