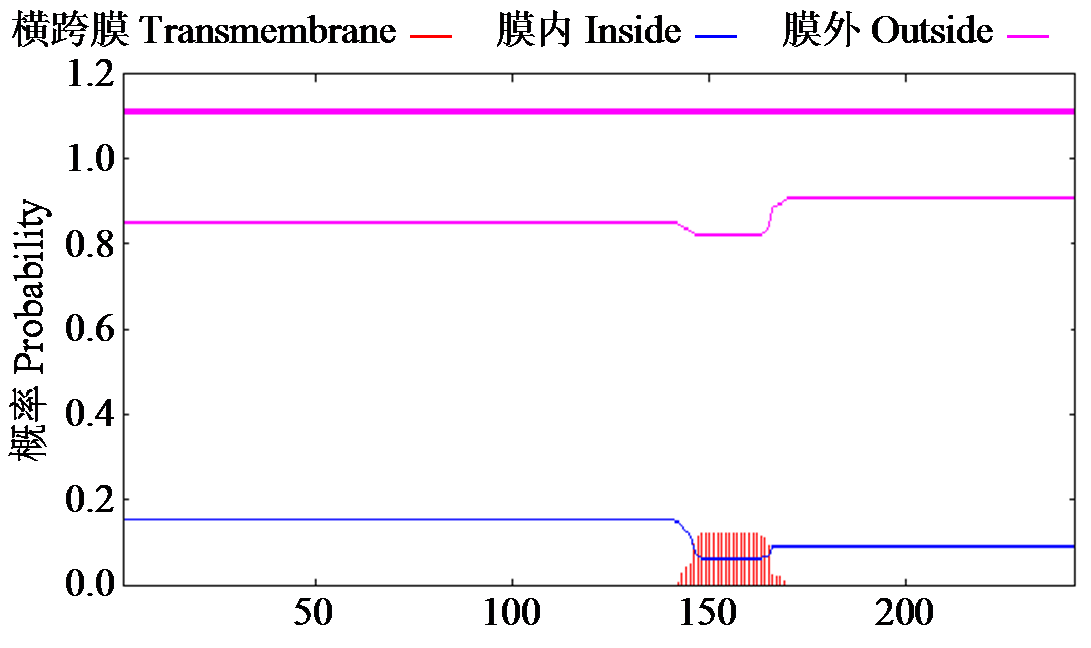
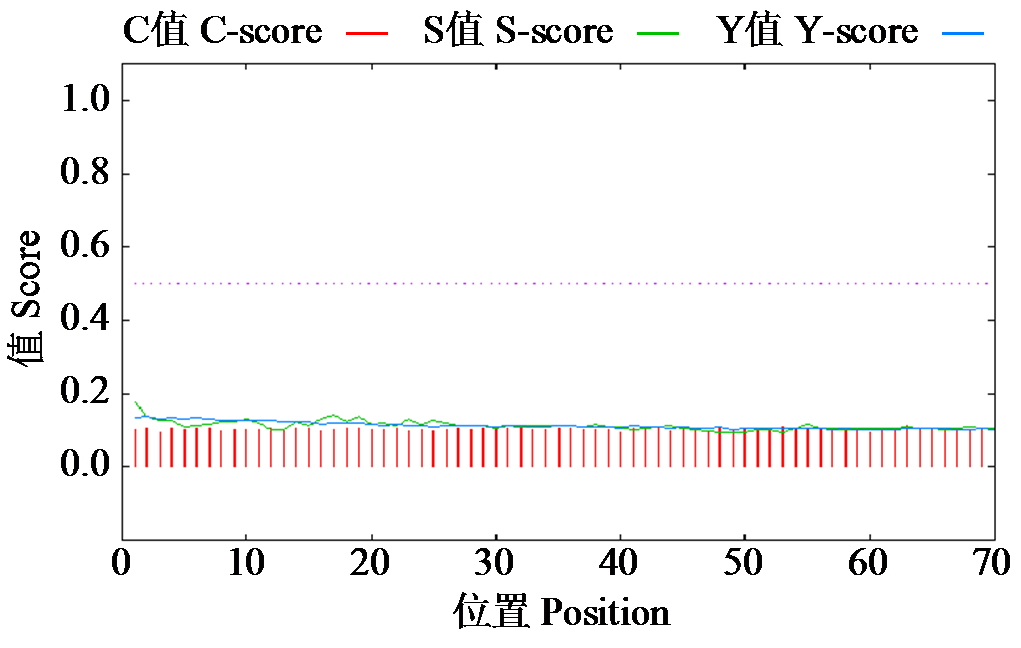
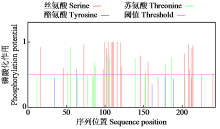
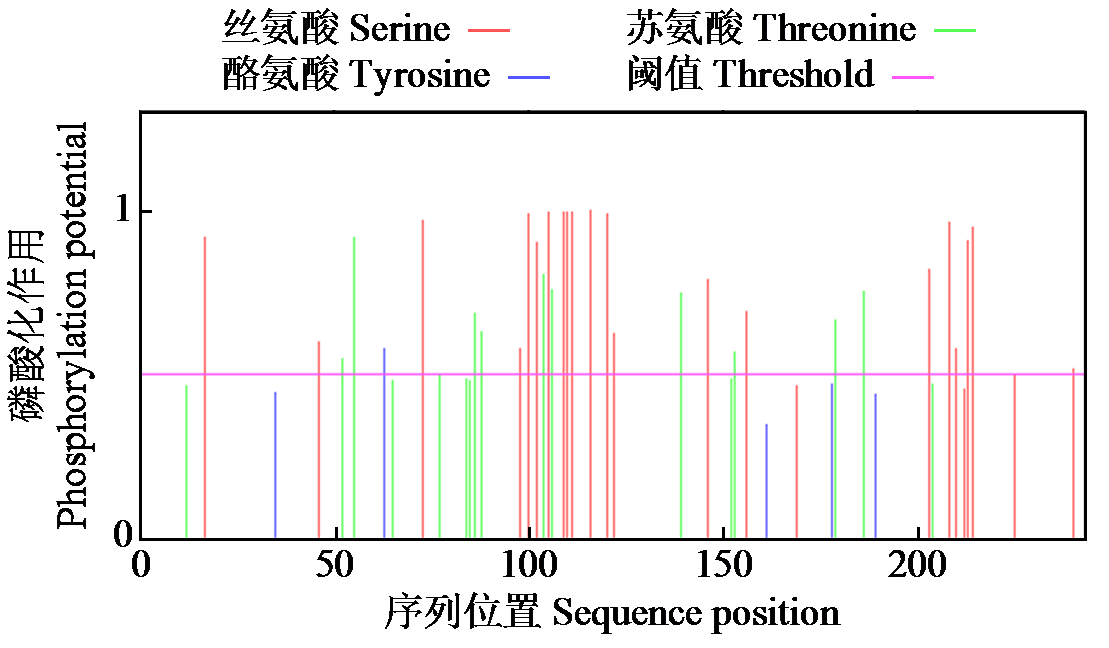
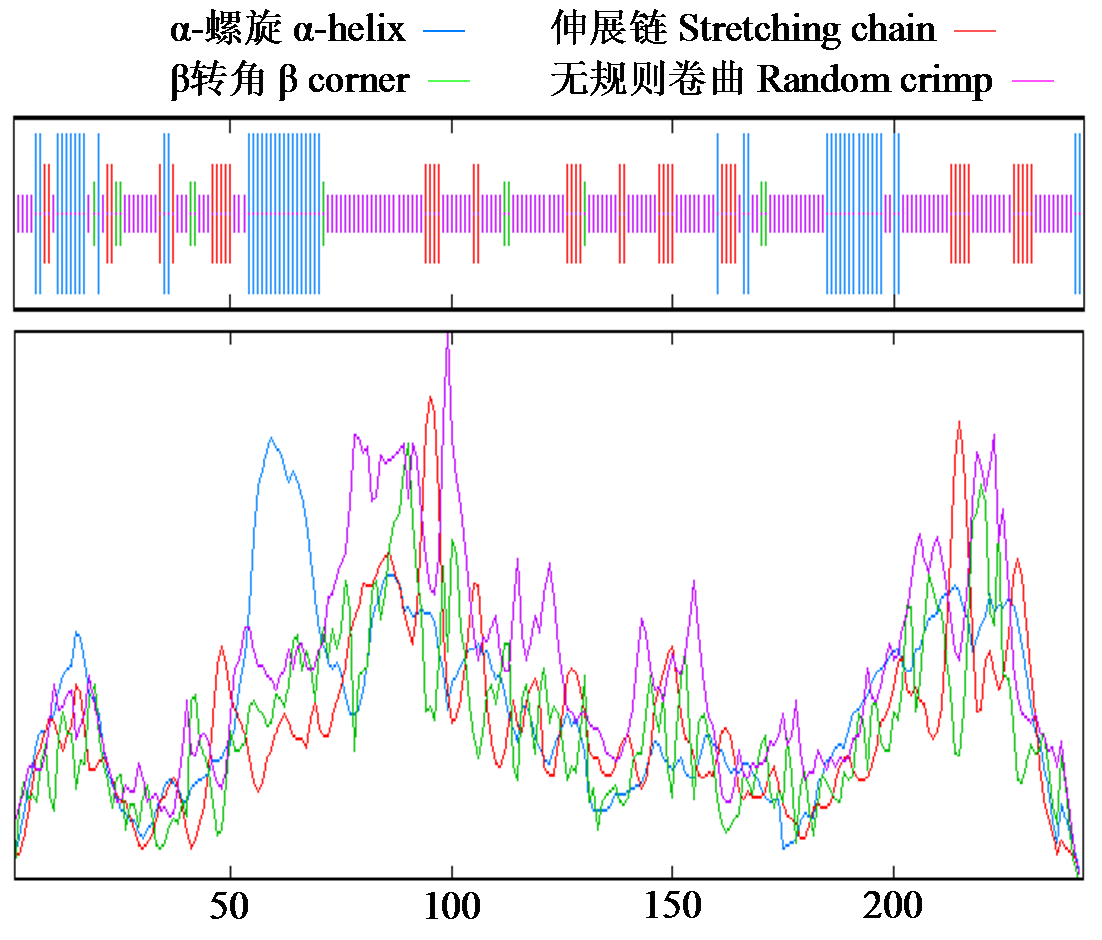
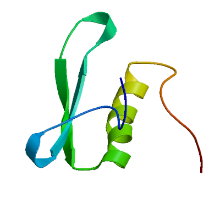
Crops ›› 2024, Vol. 40 ›› Issue (2): 23-29.doi: 10.16035/j.issn.1001-7283.2024.02.004
Previous Articles Next Articles
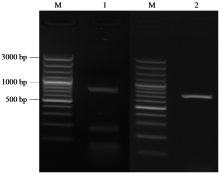
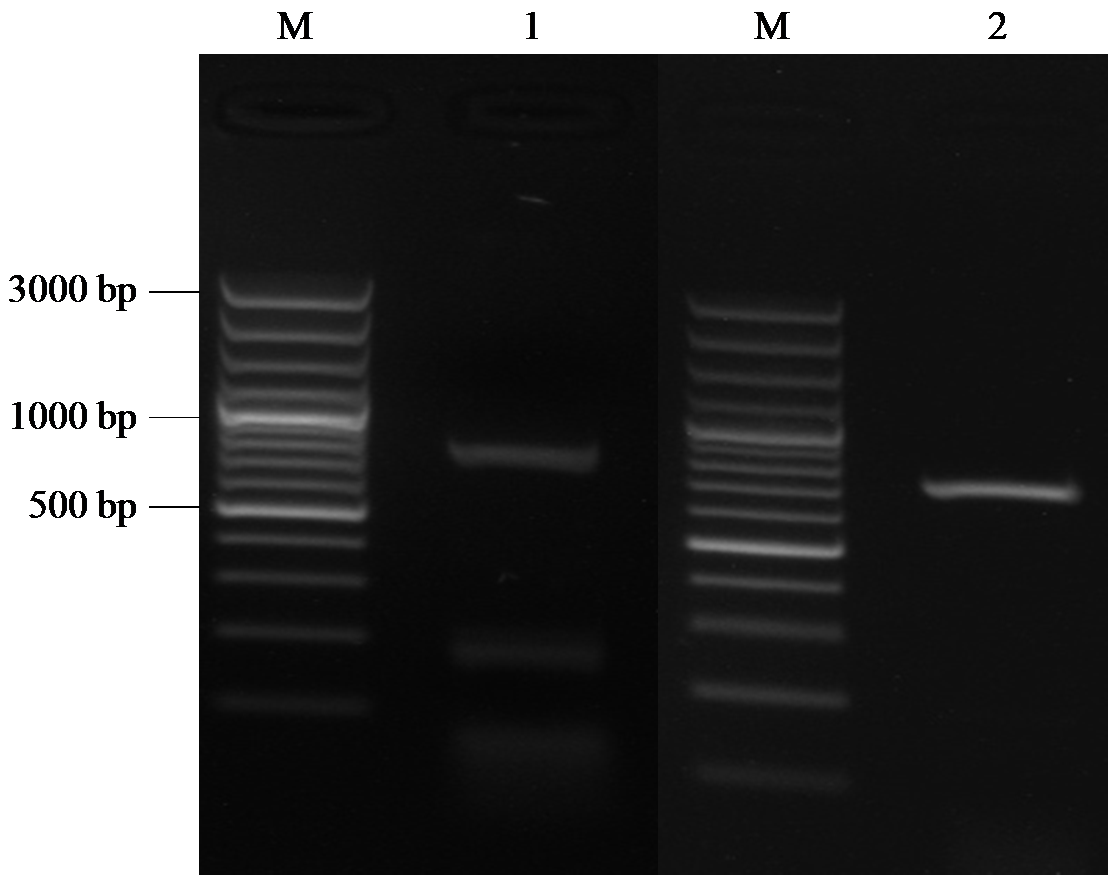
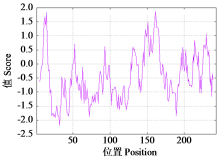
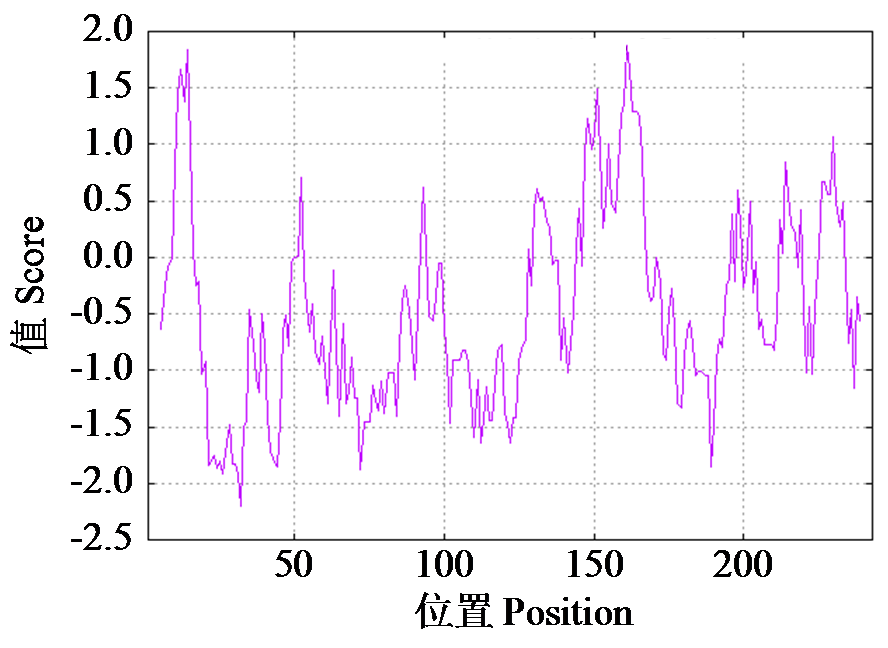
Cloning, Bioinformatics and Expression Analysis of FtERF Gene in Fagopyrum tataricum
Zhang Jun1( ), Cai Suyun1, Xu Zihao1, Hou Lei1, He Runli1(
), Cai Suyun1, Xu Zihao1, Hou Lei1, He Runli1( ), Yin Guifang2, Wang Lihua2(
), Yin Guifang2, Wang Lihua2( ), Wang Yanqing2, Lu Wenjie2, Sun Daowang2
), Wang Yanqing2, Lu Wenjie2, Sun Daowang2
- 1College of Traditional Chinese Medicine and Food Engineering, Shanxi University of Traditional Chinese Medicine, Taiyuan 030619, Shanxi, China
2Biotechnology and Genetic Germplasm Resources Research Institute, Yunnan Academy of Agricultural Sciences / Yunnan Provincial Key Laboratory of Agricultural Biotechnology / Key Laboratory of Southwestern Crop Gene Resources and Germplasm Innovation, Ministry of Agriculture and Rural Affairs, Kunming 650201, Yunnan, China
| [1] |
张强, 李艳琴. 基于矿质元素的苦荞产地判别研究. 中国农业科学, 2011, 44(22):4653-4659.
doi: 10.3864/j.issn.0578-1752.2011.22.012 |
| [2] | 张强, 李艳琴. 苦荞功能成分及其开发利用. 山西师范大学学报: 自然科学, 2009, 23(4):85-89. |
| [3] | 谭平艳, 郭皖北. 苦荞黄酮的生理功能及其作用机制的研究进展. 医学综述, 2018, 24(8):1627-1632. |
| [4] | 贾冬英, 姚开, 张海均. 苦荞麦的营养与功能成分研究进展. 粮食与饲料工业, 2012, 12(5):25-27. |
| [5] |
Liu M, Fu Q, Ma Z, et al. Genome-wide investigation of the MADS gene family and dehulling genes in tartary buckwheat (Fagopyrum tataricum). Planta, 2019, 249(5):1301-1318.
doi: 10.1007/s00425-019-03089-3 pmid: 30617544 |
| [6] | 喻武鹃. 薄壳苦荞种子代谢组学分析与AGL11基因克隆和表达研究. 贵阳: 贵州师范大学, 2020. |
| [7] |
Nakano T, Suzuki K, Fujimura T, et al. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology, 2006, 140(2):411-432.
doi: 10.1104/pp.105.073783 pmid: 16407444 |
| [8] |
张麒, 陈静, 李俐, 等. 植物AP2/ERF转录因子家族的研究进展. 生物技术通报, 2018, 34(8):1-7.
doi: 10.13560/j.cnki.biotech.bull.1985.2017-1142 |
| [9] |
Zhang J, Yin X R, Li H, et al. Ethylene Response FACTOR39- MYB 8 complex regulates low-temperature-induced lignification of loquat fruit. Journal of Experimental Botany, 2020, 71(10):3172-3184.
doi: 10.1093/jxb/eraa085 pmid: 32072171 |
| [10] |
Hu Y N, Sun Y L, Han Z Y, et al. ERF4 affects fruit firmness through TPL4 by reducing ethylene production. The Plant Journal, 2020, 103(3):937-950.
doi: 10.1111/tpj.14884 pmid: 32564488 |
| [11] |
Hu Y N, Sun H L, Han Z Y, et al. ERF4 affects fruit ripening by acting as a JAZ interactor between ethylene and jasmonic acid hormone signaling pathways. Horticultural Plant Journal, 2022, 8(6):689-699.
doi: 10.1016/j.hpj.2022.01.002 |
| [12] |
Ma R F, Xiao Y, Lv Z Y, et al. AP2/ERF transcription factor,Ii049,positively regulates lignan biosynthesis in Isatis indigotica through activating salicylic acid signaling and lignan/lignin pathway genes. Frontiers in Plant Science, 2017, 8:1361.
doi: 10.3389/fpls.2017.01361 |
| [13] |
Duan Y, Yin G F, He R L, et al. Identification of candidate genes for easily-shelled traits in Tartary buckwheat based on BSA-Seq and RNA-Seq methods. Euphytica, 2022, 218(7):1-14.
doi: 10.1007/s10681-021-02955-0 |
| [14] |
Yang Z, Tian L N, Latoszek-Green M, et al. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Molecular Biology, 2005, 58(4):585-596.
doi: 10.1007/s11103-005-7294-5 |
| [15] |
Fu C C, Han Y C, Qi X Y, et al. Papaya CpERF9 acts as a transcriptional repressor of cell-wall-modifying genes CpPME1/2 and CpPG5 involved in fruit ripening. Plant Cell Reports, 2016, 35(11):2341-2352.
doi: 10.1007/s00299-016-2038-3 |
| [16] |
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 2012, 1819(2):86-96.
doi: 10.1016/j.bbagrm.2011.08.004 |
| [17] | 吴朝昕. 薄壳苦荞果壳结构及其发育中的转录组学分析. 贵阳: 贵州师范大学, 2020. |
| [18] |
宋康华, 贾志伟, 常金梅, 等. 低温下乙烯对采后菜薹木质化及相关基因表达的影响. 园艺学报, 2019, 46(4):775-783.
doi: 10.16420/j.issn.0513-353x.2018-0561 |
| [19] |
Cai C, Chen K S, Xu W P, et al. Effect of 1-MCP on postharvest quality of loquat fruit. Postharvest Biology and Technology, 2006, 40(2):155-162.
doi: 10.1016/j.postharvbio.2005.12.014 |
| [20] |
Wang P, Zhang B, Li X, et al. Ethylene signal transduction elements involved in chilling injury in non-climacteric loquat fruit. Journal of Experimental Botany, 2010, 61(1):179-190.
doi: 10.1093/jxb/erp302 pmid: 19884229 |
| [21] |
Wang C H, Xin M, Zhou X Y, et al. The novel ethylene- responsive factor CsERF025 affects the development of fruit bending in cucumber. Plant Molecular Biology, 2017, 95(4):519-531.
doi: 10.1007/s11103-017-0671-z |
| [22] |
Zeng J K, Li X, Xu Q, et al. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnology Journal, 2015, 13(9):1325-1334.
doi: 10.1111/pbi.2015.13.issue-9 |
| [1] | Yang Enze, Xie Rui, Han Ping'an, Zhang Yonghu, Liu Jinchuan, Niu Suqing, Wen Rui, Wang Chunyong, Jin Xiaolei. Genetic Diversity and Comprehensive Evaluation of Phenotypic Traits of 162 Tartary Buckwheat Resources in Inner Mongolia [J]. Crops, 2024, 40(2): 15-22. |
| [2] | Zhang Qian, Ren Wen, Zhao Bingbing, Zhou Miaoyi, Li Hanshuai, Liu Ya, Du Hewei. Cloning and Bioinformatics Analysis of ZmMAPKKK21 Gene in Maize [J]. Crops, 2024, 40(2): 30-39. |
| [3] | Du Hanmei, Tan Lu, Chen Bo, Yu Qiuzhu, Wu Dandan, Wang Anhu. Comprehensive Evaluation of Cadmium Tolerance of Tartary Buckwheat at Seedling Stage [J]. Crops, 2024, 40(2): 40-53. |
| [4] | Zhang Yu, Yang Wenjing, Liu Xuan, Nie Fengjie, Zhang Li, Shi Lei, Zhang Guohui, Guo Zhiqian, Gong Lei. Cloning and Expression Analysis of Potato StCWIN1 Gene Promoter and Its Role under Drought Stress [J]. Crops, 2024, 40(2): 54-61. |
| [5] | Hao Yani, Pei Hongbin, Gao Zhenfeng, Zhang Yijun, Yang Zhenping. Effects of Bacillus vallismortis and Straw Replacing Phosphorus Fertilizer on Growth, Yield and Quality of Tartary Buckwheat [J]. Crops, 2024, 40(1): 204-213. |
| [6] | Lü Baolian, Yang Yuxin, Cui Licao, Shi Feng, Ma Liang, Kong Xiuying, Zhang Lichao, Ni Zhiyong. Identification of bHLH Family Transcription Factors of Wheat and Expression Analysis under Salt Stress [J]. Crops, 2024, 40(1): 65-72. |
| [7] | Wu Ying, Hu Die, Li Ting, Duan Qianyuan, Wei Ningning, Zhang Xinghua, Xu Shutu, Xue Jiquan. Analysis of WRKY Transcription Factor IIc Subfamily in Maize and Its Expression Profile under Drought [J]. Crops, 2024, 40(1): 80-89. |
| [8] | Chen Yuanyuan, Li Guangsheng, Liu Yang, He Yuqi, Zhou Meiliang, Fang Zhengwu. Molecular Cloning and Functional Identification of Resistance Gene FtTIR of Tartary Buckwheat to Blight [J]. Crops, 2023, 39(4): 44-51. |
| [9] | Zhao Pengpeng, Li Luhua, Ren Mingjian, An Chang, Hong Dingli, Li Xin, Xu Ruhong. Bioinformatics and Expression Analysis of GzCIPK7-5B Gene in Wheat [J]. Crops, 2023, 39(4): 77-84. |
| [10] | Qiu Kaihua, Fang Shumei, Liang Xilong. Functional Analysis of SRRM1-Like Transcription Factor of Magnaporthe grisea [J]. Crops, 2023, 39(3): 246-253. |
| [11] | Bai Kaihong, Abie Xiaobing, Xu Xiaoli, Jiang Na, Li Jianqiang, Luo Laixin. Analysis of Fungal Diversity in Seeds of Tartary Buckwheat from Liangshan, Sichuan Province [J]. Crops, 2023, 39(3): 260-266. |
| [12] | Li Guangsheng, Lu Xiang, Lai Dili, Zhang Kaixuan, Wang Haihua, Zhou Meiliang. Molecular Cloning and Functional Analysis of Resistance Gene FtABCG12 of Tartary Buckwheat to Blight [J]. Crops, 2023, 39(3): 43-50. |
| [13] | Meng Yaxuan, Yao Xuhang, Sun Yingqi, Zhao Xinyue, Wang Fengxia, Weng Qiaoyun, Liu Yinghui. Identification and Bioinformatics Analysis of DGAT Gene Family in Cereal Crops [J]. Crops, 2023, 39(1): 20-29. |
| [14] | Wang Junzhen, Zhou Meiliang, Li Faliang, Zhang Kaixuan, Zhu Jianfeng, Shen A’yi, Luogu Youfu, Yao Juhong, Yin Yuanjie, Wu Dongming, Zhang Jie. Breeding and Cultivation Technology of New Tartary Buckwheat Variety “Chuanqiao 6” [J]. Crops, 2022, 38(6): 241-244. |
| [15] | Zhou Fei. Bioinformatics and Expression Analysis of HaLACS7 Gene in Sunflower [J]. Crops, 2022, 38(3): 104-108. |
|
||