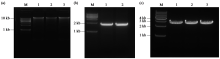
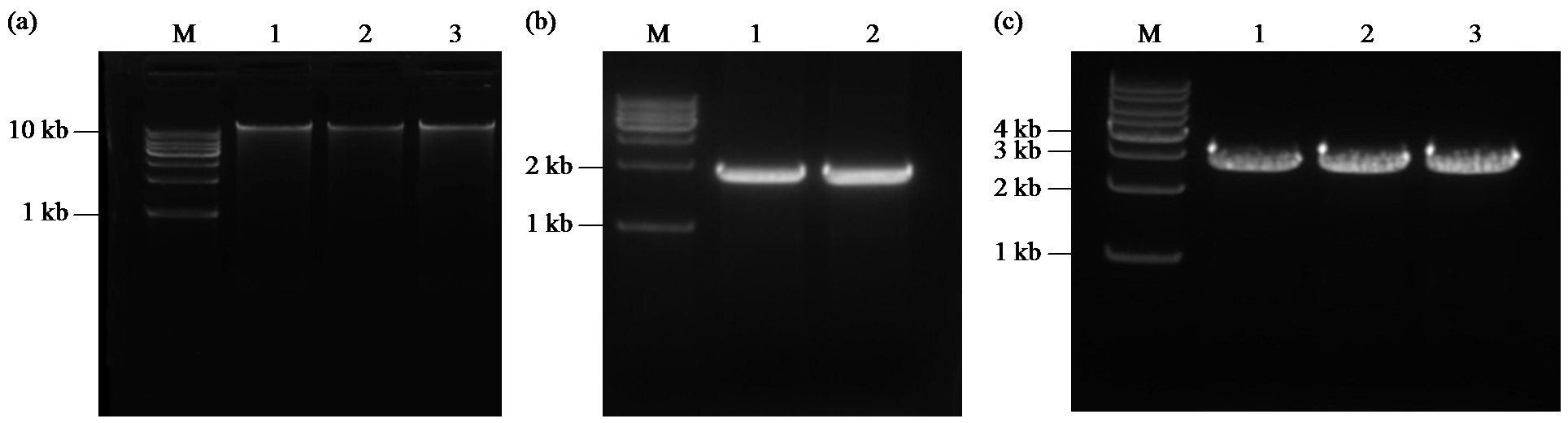
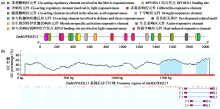
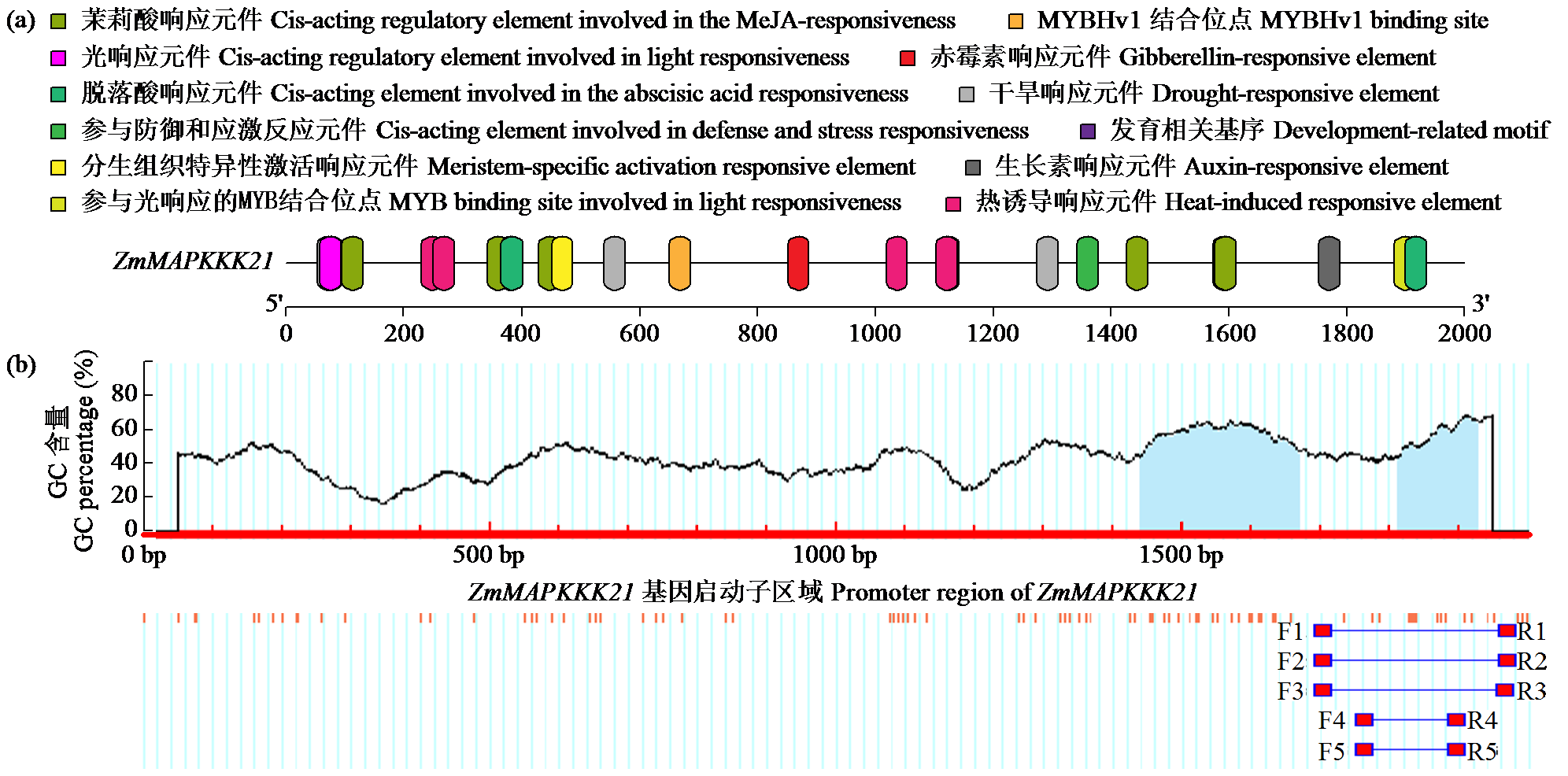
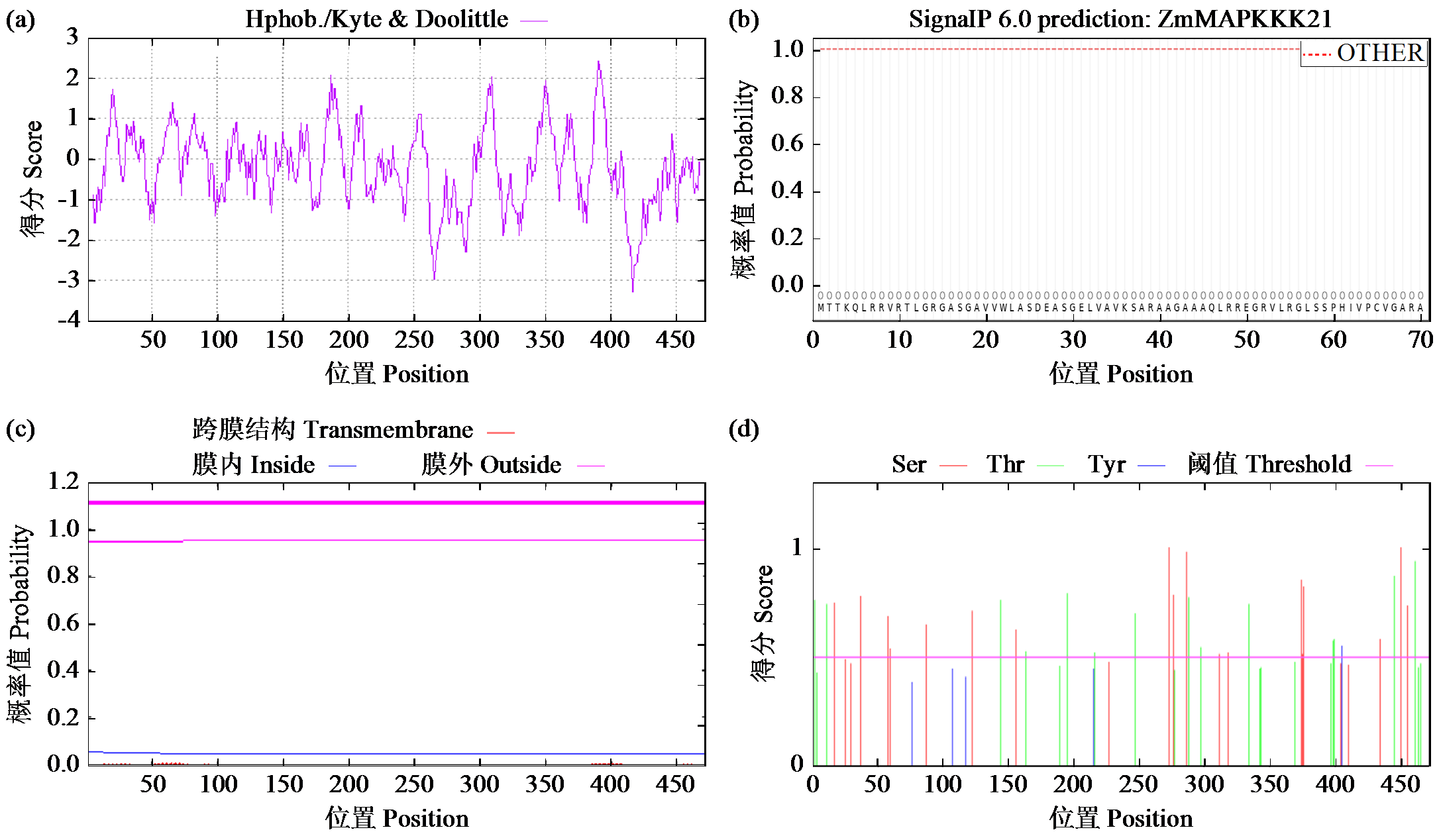
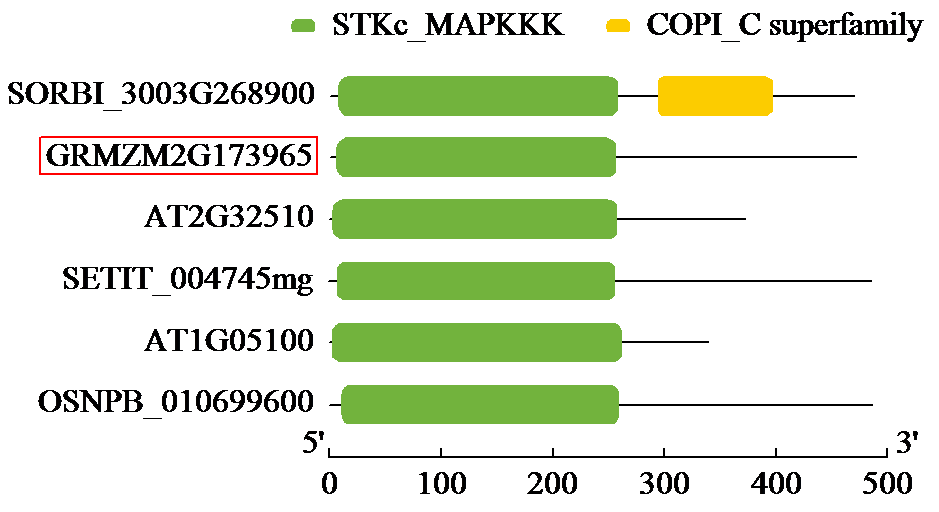
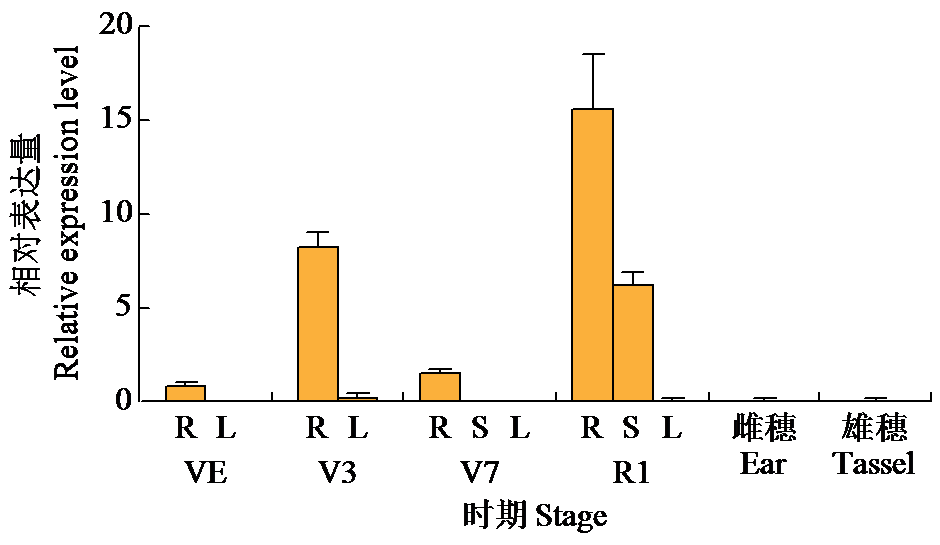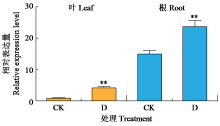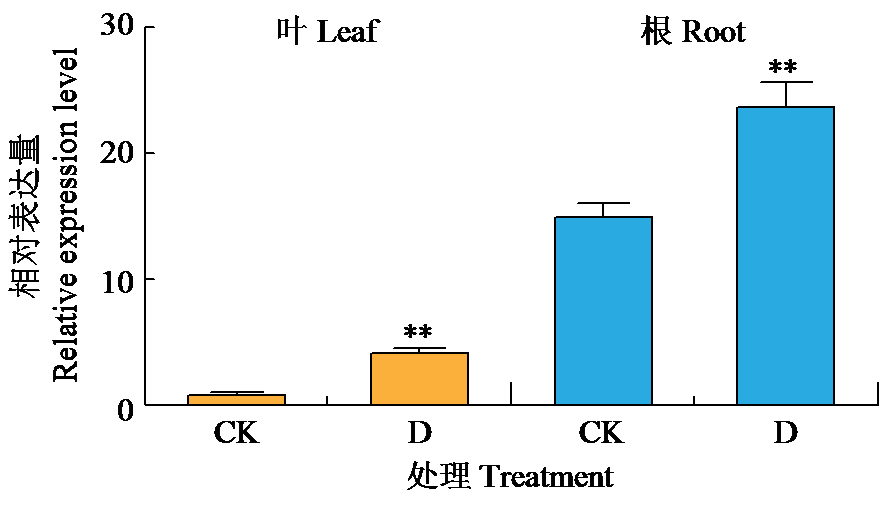Crops ›› 2024, Vol. 40 ›› Issue (2): 30-39.doi: 10.16035/j.issn.1001-7283.2024.02.005
Previous Articles Next Articles
Cloning and Bioinformatics Analysis of ZmMAPKKK21 Gene in Maize
Zhang Qian1,2( ), Ren Wen2, Zhao Bingbing2, Zhou Miaoyi2, Li Hanshuai2, Liu Ya2(
), Ren Wen2, Zhao Bingbing2, Zhou Miaoyi2, Li Hanshuai2, Liu Ya2( ), Du Hewei1(
), Du Hewei1( )
)
- 1College of Life Science, Yangtze University, Jingzhou 434025, Hubei, China
2Maize Research Institute, Beijing Academy of Agriculture & Forestry Sciences / Beijing Key Laboratory of Maize DNA Fingerprinting and Molecular Breeding, Beijing 100097, China
| [1] |
周秒依, 任雯, 赵冰兵, 等. 植物MAPK级联途径应答的非生物胁迫研究进展. 中国农业科技导报, 2020, 22(2):22-29.
doi: 10.13304/j.nykjdb.2019.0737 |
| [2] |
Zhang M M, Zhang S Q. Mitogen-activated protein kinase cascades in plant signaling. Journal of Integrative Plant Biology, 2022, 64(2):301-341.
doi: 10.1111/jipb.13215 |
| [3] |
Xu J, Zhang S Q. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends in Plant Science, 2015, 20(1):56-64.
doi: 10.1016/j.tplants.2014.10.001 pmid: 25457109 |
| [4] |
Zhang M M, Su J B, Zhang Y, et al. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Current Opinion in Plant Biology, 2018, 45:1-10.
doi: S1369-5266(17)30213-3 pmid: 29753266 |
| [5] |
Liu D D, Zhu M, Hao L L, et al. GhMAPKKK49, a novel cotton (Gossypium hirsutum L.) MAPKKK gene, is involved in diverse stress responses. Acta Physiologiae Plantarum, 2016, 38(1):13.
doi: 10.1007/s11738-015-2029-y |
| [6] |
Jonak C, Okresz L, Bogre L, et al. Complexity,cross talk and integration of plant MAP kinase signalling. Current Opinion in Plant Biology, 2002, 5(5):415-424.
doi: 10.1016/S1369-5266(02)00285-6 |
| [7] |
Rao K P, Richa T, Kumar K, et al. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Research, 2010, 17(3):139-153.
doi: 10.1093/dnares/dsq011 pmid: 20395279 |
| [8] | 刘晨, 曹小汉, 殷丹丹, 等. MAPK信号通路调控植物响应非生物胁迫的研究进展. 安徽农业科学, 2022, 50(18):9-16. |
| [9] |
Moustafa K. Improving plant stress tolerance: potential applications of engineered MAPK cascades. Trends in Biotechnology, 2014, 32(8):389-390.
doi: 10.1016/j.tibtech.2014.06.005 pmid: 24986255 |
| [10] |
Zhen W, Song Y, Ren W C, et al. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families in Fagopyrum tataricum and analysis of their expression patterns under abiotic stress. Frontiers in Genetics, 2022, 13:894048.
doi: 10.3389/fgene.2022.894048 |
| [11] |
Wang N, Liu Y S, Dong C H, et al. MdMAPKKK1 regulates apple resistance to Botryosphaeria dothidea by interacting with MdBSK1. International Journal of Molecular Sciences, 2022, 23(8):4415.
doi: 10.3390/ijms23084415 |
| [12] |
Li Y Y, Cai H X, Liu P, et al. Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochemical and Biophysical Research Communications, 2017, 484(2):292-297.
doi: 10.1016/j.bbrc.2017.01.104 |
| [13] |
Zhou M Y, Zhao B B, Han S L, et al. Comprehensive analysis of MAPK cascade genes in sorghum (Sorghum bicolor L.) reveals SbMPK14 as a potential target for drought sensitivity regulation. Genomics, 2022, 114(2):110311.
doi: 10.1016/j.ygeno.2022.110311 |
| [14] |
Daryanto S, Wang L, Jacinthe P A. Global synthesis of drought effects on maize and wheat production. PLoS ONE, 2017, 11(5):e0156362.
doi: 10.1371/journal.pone.0156362 |
| [15] |
Rai M K, Kalia R K, Singh R, et al. Developing stress tolerant plants through in vitro selection—An overview of the recent progress. Environmental and Experimental Botany, 2011, 71(1):89-98.
doi: 10.1016/j.envexpbot.2010.10.021 |
| [16] |
Liu Y, Zhou M Y, Gao Z X, et al. RNA-Seq analysis reveals MAPKKK family members related to drought tolerance in maize. PLoS ONE, 2015, 10(11):e0143128.
doi: 10.1371/journal.pone.0143128 |
| [17] |
Zhang M Y, Pan J W, Kong X D, et al. ZmMKK3, a novel maize group B mitogen-activated protein kinase kinase gene, mediates osmotic stress and ABA signal responses. Journal of Plant Physiology, 2012, 169(15):1501-1510.
doi: 10.1016/j.jplph.2012.06.008 pmid: 22835533 |
| [18] |
Zhang Z B, Li X L, Yu R, et al. Isolation,structural analysis, and expression characteristics of the maize TIFY gene family. Molecular Genetics and Genomics, 2015, 290(5):1849-1858.
doi: 10.1007/s00438-015-1042-6 |
| [19] |
Lin L, Wu J, Jiang M, et al. Plant mitogen-activated protein kinase cascades in environmental stresses. International Journal of Molecular Sciences, 2021, 22(4):1543.
doi: 10.3390/ijms22041543 |
| [20] |
Komis G, Šamajov O, Ovečka M, et al. Cell and developmental biology of plant mitogen-activated protein kinases. Annual Review of Plant Biology, 2018, 69(1):237-265.
doi: 10.1146/arplant.2018.69.issue-1 |
| [21] |
Zi S, Zhao B B, Wei S, et al. Genome-wide identification and characterization of the MAPKKK, MKK, and MPK families in Chinese elite maize inbred line Huangzaosi. The Plant Genome, 2022, 15(3):e20216.
doi: 10.1002/tpg2.v15.3 |
| [22] | 卢峰, 张飞, 段有厚. 干旱胁迫对高粱苗期物质生产及生理特性的影响. 作物杂志, 2015(2):149-153. |
| [23] | 于国红, 刘朋程, 郝洪波, 等. 不同基因型谷子对干旱胁迫的调控机制. 植物营养与肥料学报, 2022, 28(1):157-167. |
| [24] |
Wang M, Yue H, Feng K W, et al. Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.). BMC Genomics, 2016, 17(1):668.
doi: 10.1186/s12864-016-2993-7 |
| [25] |
Wu J, Wang J, Pan C T, et al. Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS ONE, 2014, 9(7):e103032.
doi: 10.1371/journal.pone.0103032 |
| [26] | 李媛媛. AtMAPKKK18调节干旱胁迫抗性的分子机理研究. 泰安: 山东农业大学, 2016. |
| [27] | 王芳, 彭云玲, 方永丰, 等. 花后干旱胁迫对不同持绿型玉米叶片衰老的影响. 水土保持通报, 2018, 38(4):60-66. |
| [28] | 陆兰姣, 王治红. 探讨玉米自交系苗期耐旱性差异分析. 农业与技术, 2016, 36(2):7,29. |
| [29] | 许璐璐, 王涵, 高盼盼, 等. 环境胁迫对植物根系形态的影响. 安徽农业科学, 2020, 48(14):16-19. |
| [1] | Wang Huaiping, Yang Mingda, Zhang Suyu, Li Shuai, Guan Xiaokang, Wang Tongchao. Effects of Different Water-Saving Irrigation Modes on Growth, Yield, and Water Utilization of Summer Maize [J]. Crops, 2024, 40(2): 206-212. |
| [2] | Zhang Jun, Cai Suyun, Xu Zihao, Hou Lei, He Runli, Yin Guifang, Wang Lihua, Wang Yanqing, Lu Wenjie, Sun Daowang. Cloning, Bioinformatics and Expression Analysis of FtERF Gene in Fagopyrum tataricum [J]. Crops, 2024, 40(2): 23-29. |
| [3] | Zhang Yu, Yang Wenjing, Liu Xuan, Nie Fengjie, Zhang Li, Shi Lei, Zhang Guohui, Guo Zhiqian, Gong Lei. Cloning and Expression Analysis of Potato StCWIN1 Gene Promoter and Its Role under Drought Stress [J]. Crops, 2024, 40(2): 54-61. |
| [4] | Hu Haochi, Wang Fugui, Zhu Kongyan, Hu Shuping, Wang Meng, Wang Zhigang, Sun Jiying, Yu Xiaofang, Bao Haizhu, Gao Julin. Effects of Straw Returning Years and Phosphorus Application on Root Growth and Yield of Maize [J]. Crops, 2024, 40(2): 80-88. |
| [5] | Feng Yong, Hou Xuguang, Xue Chunlei, Zhang Laihou, Song Guodong, Su Minli, Fu Xiaohua, Sun Yuyan. Division of Suitable Ecological Regions of Maize Varieties in Inner Mongolia [J]. Crops, 2024, 40(1): 23-30. |
| [6] | Wang Haitao, Ren Chunmei, Dong Yan, Li Shuo, Cheng Zhaobang, Ji Yinghua. Molecular Detection and Identification of Maize Yellow Mosaic Virus on Sorghum in Huai’an, Jiangsu [J]. Crops, 2024, 40(1): 233-238. |
| [7] | Ma Juan, Huang Lu, Yu Ting, Guo Guojun, Zhu Weihong, Liu Jingbao. Multi-Locus Genome-Wide Association Study and Genomic Prediction for General Combining Ability of Maize Ear Diameter [J]. Crops, 2024, 40(1): 31-39. |
| [8] | Lü Baolian, Yang Yuxin, Cui Licao, Shi Feng, Ma Liang, Kong Xiuying, Zhang Lichao, Ni Zhiyong. Identification of bHLH Family Transcription Factors of Wheat and Expression Analysis under Salt Stress [J]. Crops, 2024, 40(1): 65-72. |
| [9] | Wu Ying, Hu Die, Li Ting, Duan Qianyuan, Wei Ningning, Zhang Xinghua, Xu Shutu, Xue Jiquan. Analysis of WRKY Transcription Factor IIc Subfamily in Maize and Its Expression Profile under Drought [J]. Crops, 2024, 40(1): 80-89. |
| [10] | Jin Yu, Guo Xinyu, Zhang Ying, Li Dazhuang, Wang Jinglu. Stomatal Phenotypic Identification and Research Progress in Maize Leaves [J]. Crops, 2023, 39(6): 1-10. |
| [11] | Wu Qi, Ming Bo, Gao Shang, Yang Hongye, Zhang Chuan, Chu Zhendong, Li Shaokun. Research on the Construction Strategy of Maize Grain Dehydration Model in Cold Northeast China [J]. Crops, 2023, 39(6): 108-113. |
| [12] | Liang Zhongyu, Xue Jun, Zhang Guoqiang, Ming Bo, Shen Dongping, Fang Liang, Zhou Linli, Zhang Yuqin, Yang Hengshan, Wang Keru, Li Shaokun. Effects of Phosphorus Application Rate on Lodging Resistance of Maize under Integrated Water and Fertilizer [J]. Crops, 2023, 39(6): 190-194. |
| [13] | Cao Qingjun, Li Gang, Yang Hao, Lou Yuyong, Yang Fentuan, Kong Fanli, Li Xinbei, Zhao Xinkai, Jiang Xiaoli. The Effects of Different Tillage Practices on Seedbed Quality and Its Relationships with Seedling Population Construction and Grain Yield of Spring Maize [J]. Crops, 2023, 39(5): 249-254. |
| [14] | Yu Le, Li Lin, Huang Hongjuan, Huang Zhaofeng, Zhu Wenda, Wei Shouhui. Weed Species Composition and Community Characterization in Maize Fields in Hubei Province [J]. Crops, 2023, 39(5): 272-279. |
| [15] | Yang Zongying, Xiao Gui, Zhang Hongwei. Whole-Genome Predictive Analysis of Fresh Weight per Plant Using the Maize F1 Population [J]. Crops, 2023, 39(5): 43-48. |
|
||