Crops ›› 2022, Vol. 38 ›› Issue (3): 104-108.doi: 10.16035/j.issn.1001-7283.2022.03.015
Previous Articles Next Articles
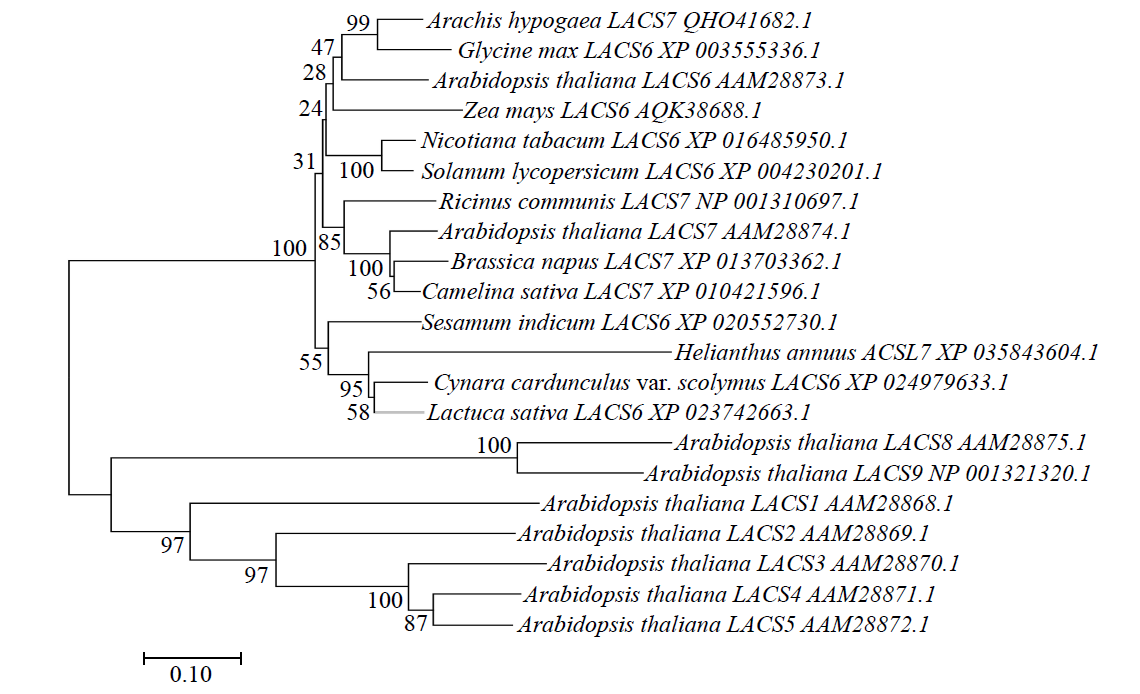
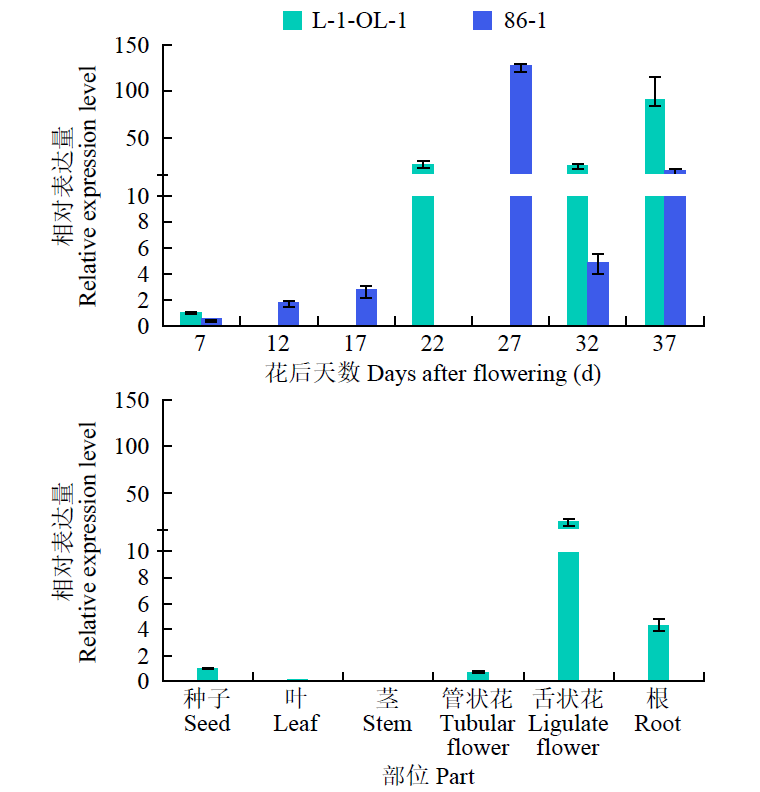
Bioinformatics and Expression Analysis of HaLACS7 Gene in Sunflower
- 1Heilongjiang Academy of Agricultural Sciences Postdoctoral Programme, Harbin 150086, Heilongjiang, China
2Industrial Crops Institute, Heilongjiang Academy of Agricultural Sciences, Harbin 150086, Heilongjiang, China
| [1] |
Jessen D, Roth C, Wiermer M, et al. Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiology, 2015, 167(2):351-366.
doi: 10.1104/pp.114.250365 pmid: 25540329 |
| [2] |
Koo A J K, Ohlrogge J B, Pollard M. On the export of fatty acids from the chloroplast. Journal of Biological Chemistry, 2004, 279(16):16101-16110.
doi: 10.1074/jbc.M311305200 |
| [3] | 汪信东, 江香梅, 章挺, 等. 樟树长链脂肪酰CoA合成酶1鉴定与功能初步分析. 分子植物育种, 2018, 16(16):105-114. |
| [4] |
Ding L N, Gu S L, Zhu F G, et al. Long-chain acyl-CoA synthetase 2 is involved in seed oil production in Brassica napus. BMC Plant Biology, 2020, 20(1):21-34.
doi: 10.1186/s12870-020-2240-x |
| [5] |
Aznar-Moreno J A. Calerón M V, Martínez-Force E, et al. Sunflower (Helianthus annuus) long-chain acyl-coenzyme A synthetases expressed at high levels in developing seeds. Physiologia Plantarum, 2014, 150(3):363-373.
doi: 10.1111/ppl.12107 |
| [6] |
Zhao L F, Haslam T M, Sonntag A, et al. Functional overlap of long-chain acyl-CoA synthetases in Arabidopsis. Plant and Cell Physiology, 2019, 60(5):1041-1054.
doi: 10.1093/pcp/pcz019 |
| [7] |
Fulda M, Shockey J, Werber M, et al. Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid β-oxidation. The Plant Journal, 2002, 32(1):93-103.
doi: 10.1046/j.1365-313X.2002.01405.x |
| [8] | 徐靖, 李辉亮, 朱家红, 等. 植物脂肪酸β-氧化的研究进展. 生物技术通讯, 2008, 19(1):141-144. |
| [9] |
Shockey J M, Fulda M, Browse J A. Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiology, 2002, 129(4):1710-1722.
pmid: 12177484 |
| [10] | 江南, 谭晓风, 张琳, 等. 油茶长链脂肪酰基CoA合成酶基因1(CoLACS1) 分子特征与表达分析. 植物遗传资源学报, 2017, 18(1):139-147. |
| [11] | 徐日荣, 陈湘瑜, 唐兆秀. 花生长链酰基辅酶A合成酶6基因(LACS6)的克隆、鉴定与组织表达. 福建农业学报, 2016, 31(11):1164-1170. |
| [12] | 汪信东, 章挺, 杨海宽, 等. 樟树长链脂肪酰基CoA合成酶基因9克隆与表达分析. 广西植物, 2018, 38(10):1335-1345. |
| [13] |
Xu Y, Holic R, Li D, et al. Substrate preferences of long-chain acyl-CoA synthetase and diacylglycerol acyltransferase contribute to enrichment of flax seed oil with а-linolenic acid. Biochemical Journal, 2018, 475(8):1473-1489.
doi: 10.1042/BCJ20170910 |
| [1] | Jia Xiuping, Mao Xuhui, Liang Gensheng, Liu Runping, Liu Feng, Wang Xingzhen. Analysis of Physiological and Biochemical Mechanism and Growth and Development Characteristics of Saline and Alkali Resistance in Sunflower [J]. Crops, 2022, 38(5): 146-152. |
| [2] | Shi Bixian, Tao Jianfei, Gao Yan, Xie Huihong, Abulimiti·Aierken , Cheng Pingshan, Maitituersun·Sadike , Sha Hong. Effects of Different Planting Densities on the Morphological Traits and Yields of Three Confectionery Sunflower Varieties [J]. Crops, 2022, 38(5): 195-200. |
| [3] | Ling Yibo, Feng Yunge, Wang Binjie, Zhang Kai, Chen Nianlai. Effects of Density and Row Spacing on Canopy Structure and Photosynthetic Characteristics in Sunflower [J]. Crops, 2022, 38(3): 155-160. |
| [4] | Guo Shuchun, Miao Hongmei, Li Suping, Yu Haifeng, Nie Hui, Mu Yingnan, Wen Xinyu, Liang Chen, Zhang Haibin, Shao Ying. Research Advances in the Breeding Study of Sunflower Resistance to Orobanche [J]. Crops, 2022, 38(3): 27-32. |
| [5] | Yang Xiaolin, Duan Ying, Cai Suyun, He Runli, Yin Guifang, Wang Yanqing, Lu Wenjie, Sun Daowang, Wang Lihua. Molecular Cloning and Bioinformatics Analyzing of Laccase in Fagopyrum tataricum [J]. Crops, 2022, 38(3): 73-79. |
| [6] | Lu Ping, Kang Qingfang, Zhao Mengyao, Zhang Fengjie, Wu Qiangqiang, Ma Fangfang, Wang Yushen, Han Yuanhuai, Wang Xingchun, Li Xueyin. Identification and Functional Analysis of miR169 Family and Its Target Genes in Setaria italica [J]. Crops, 2022, 38(2): 54-63. |
| [7] | Li Yang, Ren Xiaoci, Li Xiaowei, Li Weitang, Huang Wei, He Zhongguo, Wang Baizhong. Combining Ability and Genetic Analysis of Yield and Main Grain Characteristics of Edible Sunflower [J]. Crops, 2022, 38(2): 75-80. |
| [8] | Yin Guifang, Duan Ying, Yang Xiaolin, Cai Suyun, Wang Yanqing, Lu Wenjie, Sun Daowang, He Runli, Wang Lihua. Cloning and Bioinformatics Analysis of FtC4H Gene from Tartary Buckwheat [J]. Crops, 2022, 38(1): 77-83. |
| [9] | Zhao Xuanwei, Zhao Yajie, Tian Zhendong, Hu Shuping, Zhao Rong, Ren Yaning, Bao Haizhu, Gao Julin. Response of Dry Matter Transportion and Yield to Sowing Date and Planting Density on Sunflower [J]. Crops, 2021, 37(3): 185-189. |
| [10] | Zhou Fei, Wang Wenjun, Liu Yan, Ma Jun, Wang Jing, Wu Liren, Guan Hongjiang, Huang Xutang. Establishment of Near-Infrared Spectroscopy Model for the Contents of Fat and Fatty Acids in Sunflower Husked Seeds [J]. Crops, 2021, 37(2): 200-206. |
| [11] | Di Na, Zheng Na, Han Haijun, Wang Jing, Cui Chao, Zheng Xiqing. The Stimulatory Effect of Different Sunflower Varieties Root Exudates on Germination of Sunflower Broomrape [J]. Crops, 2020, 36(6): 197-201. |
| [12] | Yang Haifeng, Duan Xueyan, Wei Ling, Liu Bo. The Genetic Study of Yield Traits in Edible Sunflower [J]. Crops, 2020, 36(5): 93-97. |
| [13] | Shan Feibiao, Du Ruixia, Wang Yongxing, Yang Qinfang, Liu Chunhui, Chen Yang. Genetic Diversity Analysis of Sunflower (Helianthus annuus L.) Based on DUS Testing [J]. Crops, 2020, 36(4): 107-113. |
| [14] | Zhang Haiyang, Li Haiyan, Meng Qinglin, He Chaoqun, Liu Shuqing. Effects of Different Fungicides on Field Control of Sunflower Sclerotinia Rot [J]. Crops, 2020, 36(4): 202-205. |
| [15] | Zhou Zichao, Hou Jianhua, Zhen Zilong, Shi Huimin. Drought-Resistance Identification and Evaluation of 152 Sunflower Recombinant Inbred Lines (RILs) at Seedling Stage [J]. Crops, 2020, 36(3): 47-52. |
|
||