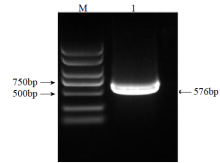
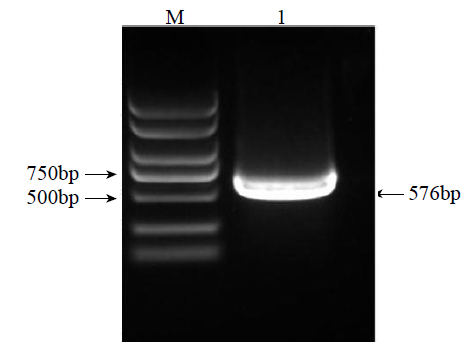
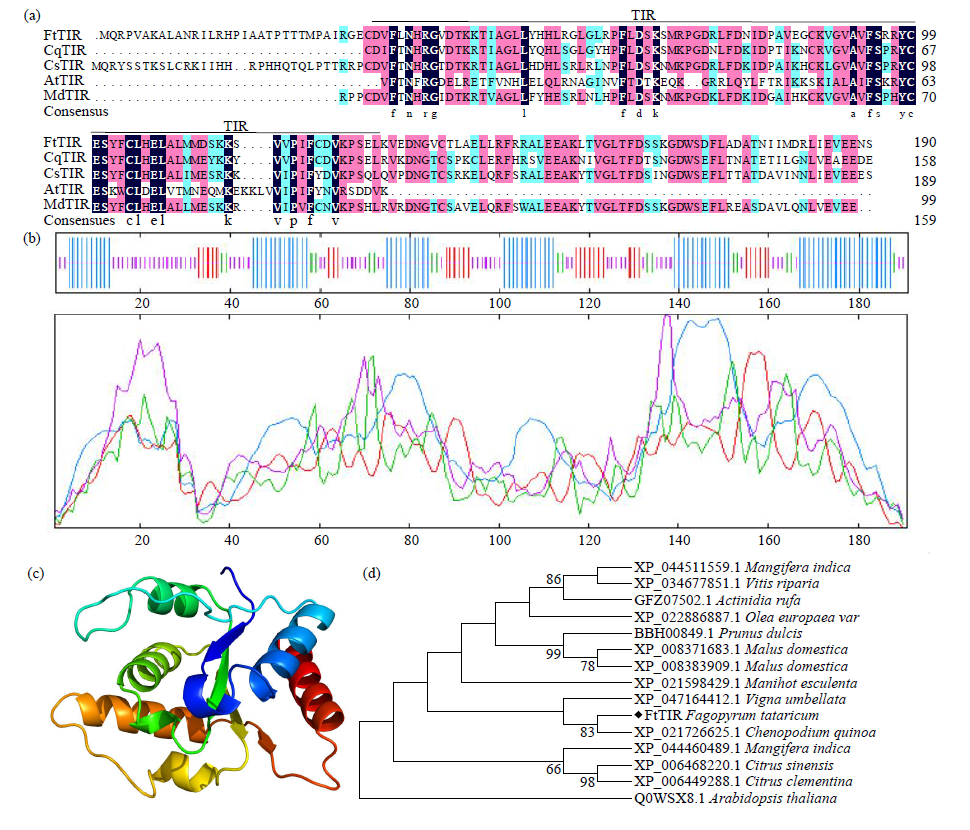
Crops ›› 2023, Vol. 39 ›› Issue (4): 44-51.doi: 10.16035/j.issn.1001-7283.2023.04.007
Previous Articles Next Articles
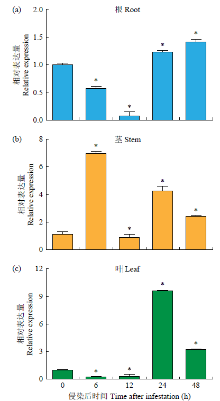
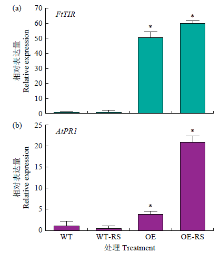
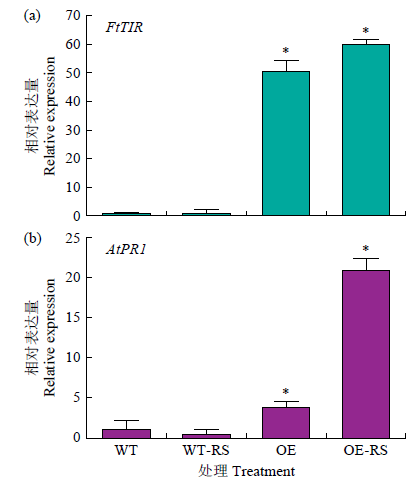
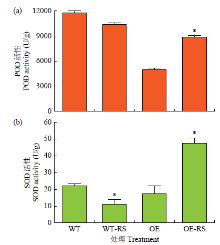
Molecular Cloning and Functional Identification of Resistance Gene FtTIR of Tartary Buckwheat to Blight
Chen Yuanyuan1,2( ), Li Guangsheng2,3, Liu Yang2,4, He Yuqi2, Zhou Meiliang2(
), Li Guangsheng2,3, Liu Yang2,4, He Yuqi2, Zhou Meiliang2( ), Fang Zhengwu1
), Fang Zhengwu1
- 1College of Agriculture, Yangtze University, Jingzhou 434025, Hubei, China
2Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
3College of Life Science and Health, Hunan University of Science and Technology, Xiangtan 411201, Hunan, China
4College of Agronomy and Biotechnology, Southwest University, Chongqing 400700, China
| [1] |
闫佳, 刘雅琼, 侯岁稳. 植物抗病蛋白研究进展. 植物学报, 2018, 53(2):250-263.
doi: 10.11983/CBB17148 |
| [2] | 高玉霞. 植物病原与抗病基因的进化研究. 南京:南京师范大学, 2018. |
| [3] | 许凤, 杨秀梅, 张丽芳, 等. 观赏植物抗病育种研究进展. 江苏农业科学, 2021, 49(7):44-51. |
| [4] | 尹玲, 方辉, 黄羽, 等. 植物TIR-NB-LRR类型抗病基因各结构域的研究进展. 广西植物, 2017, 37(2):186-190. |
| [5] | Van G C, Esmenjaud D. TNL genes in peach: insights into the post-LRR domain. BMC Genomics, 2016, 30(17):317. |
| [6] | 董亚萍, 杨仕梅, 赵德刚, 等. 烟草TIR-NBS基因家族的生物信息学分析. 广西植物, 2020, 40(6):891-900. |
| [7] |
柴亚茹, 丁一娟, 周思钰, 等. HIGS-SsCCS转基因拟南芥的菌核病抗性鉴定. 中国农业科学, 2020, 53(4):761-770.
doi: 10.3864/j.issn.0578-1752.2020.04.008 |
| [8] |
Bernoux M, Burdett H, Williams S J, et al. Comparative analysis of the flax immune receptors L6 and L 7 suggests an equilibrium-based switch activation model. The Plant Cell, 2016, 28(1):146-159.
doi: 10.1105/tpc.15.00303 pmid: 26744216 |
| [9] | Dinesh-Kumar S P, Tham W H, Baker B J.Structure-function analysis of the tobacco mosaic virus resistance gene N. Proceedings of the National Academy of Sciences of the United States of America, 2000, 97(26):14789-14794. |
| [10] |
Schreiber K J, Bentham A, Williams S J, et al. Multiple domain associations within the Arabidopsis immune receptor RPP 1 regulate the activation of programmed cell death. PLoS Pathogens, 2016, 12(7):e1005769.
doi: 10.1371/journal.ppat.1005769 |
| [11] |
Zhang Y, Dorey S, Swiderski M, et al. Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. The Plant Journal, 2004, 40(2):213-224.
doi: 10.1111/tpj.2004.40.issue-2 |
| [12] |
唐宇, 邵继荣, 周美亮. 中国荞麦属植物分类学的修订. 植物遗传资源学报, 2019, 20(3):646-653.
doi: 10.13430/j.cnki.jpgr.20181210001 |
| [13] | 罗吉. 荞麦立枯病生防菌的筛选鉴定及生防效果初探. 成都:成都大学, 2021. |
| [14] | 齐杨菊, 陈振江, 李振霞, 等. 荞麦病害研究进展. 草业科学, 2020, 37(1):75-86. |
| [15] |
卢文洁, 李春花, 王艳青, 等. 荞麦轮纹病抗性鉴定方法的建立及荞麦抗病种质资源的筛选. 中国农学通报, 2017, 33(12):98-102.
doi: 10.11924/j.issn.1000-6850.casb16120120 |
| [16] |
Gassmann W, Hinsch M E, Staskawicz B J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. The Plant Journal, 1999, 20(3):265-277.
doi: 10.1046/j.1365-313X.1999.t01-1-00600.x |
| [17] |
Subedi S, Dhami N B, Gurung S B, et al. Assessment of disease resistance and high yielding traits of common buckwheat genotypes in subtropical climate of Nepal. SAARC Journal of Agriculture, 2020, 18(1):143-152.
doi: 10.3329/sja.v18i1.48388 |
| [18] |
徐琴琴, 陈卫良, 毛碧增. 立枯丝核菌毒素的研究进展. 核农学报, 2020, 34(10):2219-2225.
doi: 10.11869/j.issn.100-8551.2020.10.2219 |
| [19] | Ade J, DeYoung B J, Golstein C, et al. Indirect activation of a plant nucleotide binding site: leucine-rich repeat protein by a bacterial protease. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(7):2531-2536. |
| [20] |
Michael W L, Swiderski M R, Li Y, et al. The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. The Plant Journal, 2006, 47(6):829-840.
doi: 10.1111/tpj.2006.47.issue-6 |
| [21] | Bernoux M, Ve T, Williams S, et al. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host & Microbe, 2011, 9(3):200-211. |
| [22] | 尹秀娟, 涂怡, 刘昱. TLR与PI3K-Akt通路交互调控的研究进展. 生物化工, 2022, 8(2):124-130. |
| [23] | Tao Y, Yuan F, Leister R T, et al. Mutational analysis of the Arabidopsis nucleotide binding site: leucine-rich repeat resistance gene RPS2. The Plant Cell, 2000, 12(12):2541-2554. |
| [24] |
Kawchuk L M, Martin R R, Mcpherson J. Resistance in transgenic potato expressing the potato leafroll virus coat protein gene. Molecular Plant-Microbe Interactions, 1990, 3(5):301-307.
doi: 10.1094/MPMI-3-301 |
| [25] |
Hwang C F, Bhakta A V, Truesdell G M, et al. Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. The Plant Cell, 2000, 12(8):1319-1329.
doi: 10.1105/tpc.12.8.1319 |
| [1] | Bai Kaihong, Abie Xiaobing, Xu Xiaoli, Jiang Na, Li Jianqiang, Luo Laixin. Analysis of Fungal Diversity in Seeds of Tartary Buckwheat from Liangshan, Sichuan Province [J]. Crops, 2023, 39(3): 260-266. |
| [2] | Li Guangsheng, Lu Xiang, Lai Dili, Zhang Kaixuan, Wang Haihua, Zhou Meiliang. Molecular Cloning and Functional Analysis of Resistance Gene FtABCG12 of Tartary Buckwheat to Blight [J]. Crops, 2023, 39(3): 43-50. |
| [3] | Wang Junzhen, Zhou Meiliang, Li Faliang, Zhang Kaixuan, Zhu Jianfeng, Shen A’yi, Luogu Youfu, Yao Juhong, Yin Yuanjie, Wu Dongming, Zhang Jie. Breeding and Cultivation Technology of New Tartary Buckwheat Variety “Chuanqiao 6” [J]. Crops, 2022, 38(6): 241-244. |
| [4] | Shi Xian, Li Hongyou, Lu Bingyue, Zhou Yun, Zhao Jiju, Zhao Mengli, Liang Jing, Meng Hengling. Physiological Responses of Three Tartary Buckwheat Varieties to Salt Stress and Evaluation of Salt Tolerance [J]. Crops, 2022, 38(3): 149-154. |
| [5] | Yin Guifang, Duan Ying, Yang Xiaolin, Cai Suyun, Wang Yanqing, Lu Wenjie, Sun Daowang, He Runli, Wang Lihua. Cloning and Bioinformatics Analysis of FtC4H Gene from Tartary Buckwheat [J]. Crops, 2022, 38(1): 77-83. |
| [6] | Weng Wenfeng, Wu Xiaofang, Zhang Kaixuan, Tang Yu, Jiang Yan, Ruan Jingjun, Zhou Meiliang. The Overexpression of FtbZIP5 Improves Accumulation of Flavonoid in the Hairy Roots of Tartary Buckwheat and Its Salt Tolerance [J]. Crops, 2021, 37(4): 1-9. |
| [7] | Jia Ruiling, Zhao Xiaoqin, Nan Ming, Chen Fu, Liu Yanming, Wei Liping, Liu Junxiu, Ma Ning. Genetic Diversity Analysis and Comprehensive Assessment of Agronomic Traits of 64 Tartary Buckwheat Germplasms [J]. Crops, 2021, 37(3): 19-27. |
| [8] | Jin Jiangang, Tian Zaifang. Grey Correlation Analysis of Introduced Tartary Buckwheat in the Northern Shanxi [J]. Crops, 2021, 37(2): 52-56. |
| [9] | Ma Mingchuan, Liu Longlong, Liu Zhang, Zhou Jianping, Nan Chenghu, Zhang Lijun. Analysis of SSR Loci in Whole Genome and Development of Molecular Markers in Tartary Buckwheat [J]. Crops, 2021, 37(1): 38-46. |
| [10] | Lu Xiaoling, He Ming, Zhang Kaixuan, Liao Zhiyong, Zhou Meiliang. Study on the Cloning and Transformation of Rhamnose Transferase FtF3GT1 Gene in Tartary Buckwheat [J]. Crops, 2020, 36(5): 33-40. |
| [11] | Yang Xuele, Zhang Lu, Li Zhiqing, He Luqiu. Diversity Analysis of Tartary Buckwheat Germplasms Based on Phenotypic Traits [J]. Crops, 2020, 36(5): 53-58. |
| [12] | Li Chunhua, Huang Jinliang, Yin Guifang, Wang Yanqing, Lu Wenjie, Sun Daowang, Wang Chunlong, Guo Laichun, Hong Bo, Ren Changzhong, Wang Lihua. Genetic Analysis of Grain Shape Related Traits in Tartary Buckwheat [J]. Crops, 2020, 36(3): 42-46. |
| [13] | Chengrui Ma,Dabing Xiang,Yan Wan,Jianyong Ouyang,Yue Song,Zhengsong Tang,Jianying Liu,Gang Zhao. Difference Analysis of Spatial Distribution Characteristics of Different Tartary Buckwheat Varieties [J]. Crops, 2020, 36(1): 35-40. |
| [14] | Yang Tian,Zhang Yongqing,Dong Fuhui,Ma Xingxing,Xue Xiaojiao. Research on the Root Growth of Different Drought-Resistant Fagopyrum tataricum under Different Water Conditions [J]. Crops, 2019, 35(6): 76-82. |
| [15] | Song Lifang,Feng Meichen,Zhang Meijun,Xiao Lujie,Wang Chao,Yang Wude,Song Xiaoyan. Effects of Exogenous Selenium on the Growth and Development of Tartary Buckwheat and Selenium Content in Grains [J]. Crops, 2019, 35(3): 150-154. |
|
||