开放科学(资源服务)标识码(OSID): 
植物油脂的合成过程主要包括质体内脂肪酸的从头合成、脂肪酸在内质网中的加工和三酰甘油(TAG)的合成积累,由多种基因协同作用完成[4]。
LEC有LEC1和LEC2两种类型[7]。其中LEC2是从拟南芥T-DNA插入突变体中发现并分离得到的[8],其属于B3转录因子超家族中的LAV家族的AFL亚家族[9]。B3结构域(植物特有的一种DNA结合基序)是一种转录激活因子,能激活胚胎发育成熟阶段贮藏蛋白编码基因的转录,同时具有调控胚对脱落酸的敏感性、胚中叶绿素降解和花青素合成、增强种子抗旱能力和抑制不成熟种子萌发等重要生理功能,这些过程均与胚和子叶的正常发育有关[10]。B3结构域编码110个氨基酸,能与核心DNA元件(CTAG)结合[11]。同属于AFL亚家族的ABI3、FUS3和LEC2的进化关系分析结果显示,LEC2基因仅存在于双子叶植物中,而在藻类、苔藓、蕨类和单子叶植物中均未发现,进一步推测最早在绿藻中分化出ABI3基因,在陆生植物中首先分化出FUS3基因,而LEC2基因最后在双子叶植物中分化产生[12]。过表达拟南芥AtLEC2基因可增加叶片脂肪酸的积累,同时提高LEC1、FUS3和ABI3基因的表达量[13]。在拟南芥中超表达蓖麻RcLEC2基因,能激活油体蛋白相关基因的表达,种子含油量增加约2.7倍[14]。在拟南芥中证实,AtLEC2通过调控下游AtWRI1基因的表达来间接调控脂肪酸合成相关基因的转录表达[15]。过表达LEC2基因可激活一些生长素响应基因的表达[16]。拟南芥LEC2蛋白可结合贮藏蛋白基因At2S3上游启动子区的RY-G-Box和RY元件(CATGCATG),进而调控某些种子特异性基因的表达[17]。拟南芥Lec2缺失突变体种子的蛋白质和油含量分别比野生型减少了15%和30%,蔗糖和淀粉含量均有所提高[18]。由此可见,LEC2在调控下游基因以及提高种子含油量方面发挥重要作用。
了解亚麻荠CsLEC2家族成员的基本理化性质、功能及进化对亚麻荠种子油脂的合成具有重要的意义。本文以十字花科拟南芥AtLEC2蛋白为参考序列,在亚麻荠数据库中进行BLAST搜索,得到3个编码CsLEC2转录因子的基因,利用生物信息学方法对这些蛋白进行了理化性质、基因结构、跨膜结构域、信号肽和亚细胞定位预测、二级结构和三级结构预测、保守结构、系统进化和启动子顺式元件分析。采用半定量与实时荧光定量检测CsLEC2基因的时空表达特性,以及通过共表达分析预测CsLEC2可能调控的下游基因,为解析CsLEC2介导亚麻荠油脂合成调控机制及后续基因修饰、提高亚麻荠等油料作物种子含油量提供参考依据。
1 材料与方法
1.1 试验材料
供试亚麻荠材料种植于山西农业大学农作站试验田。定量表达用的根、茎、叶均采集于亚麻荠8~10片真叶幼苗;花和不同时期种子分别采集于盛花期和开花后10、20、30和40d。试验材料采集后,经液氮速冻并于-80℃超低温冰箱中保存备用。
1.2 数据来源与序列鉴定
以拟南芥AtLEC2蛋白序列(NCBI登录号为NP_564304.1)为参考序列,在亚麻荠基因组数据库中进行BLASTP搜索,发现CsLEC2有3条同源蛋白序列(Csa03g031590.1、Csa14g035910.1和Csa17g039140.1),下载3条CsLEC2蛋白序列。采用保守结构域数据库(http://www.ncbi.nlm.nih.gov/Structure/cdd/docs/cdd_search.html)、SMART(http://smart.embl-heidelberg.de/)和Pfam(http://pfam.xfam.org/)鉴定上述编码蛋白的功能结构域。根据染色体分布,将3条蛋白序列分别命名为CsLEC2.1(Csa03g031590.1)、CsLEC2.2(Csa14g035910.1)和CsLEC2.3(Csa17g039140.1)。在NCBI数据库中用CsLEC2蛋白序列Protein BLAST,下载其他物种的LEC2蛋白序列(表1)。
表1 各物种的LEC2蛋白序列基本信息
Table 1
| 物种Species | 名称 Name | NCBI编号 NCBI number |
|---|---|---|
| 木薯Manihot esculenta | MeLEC2 | XP_021598372.1 |
| 麻风树Jatropha curcas | JcLEC2 | XP_012092409.1 |
| 蓖麻Ricinus communis | RcLEC2 | NP_001310615.1 |
| 毛果杨Populus trichocarpa | PtLEC2 | XP_024454872.1 |
| 银中杨Populus alba | PaLEC2 | TKR58082.1 |
| 可可Theobroma cacao | TcLEC2 | XP_017978733.1 |
| 克莱门柚Citrus clementina | CcLEC2 | XP_024046749.1 |
| 杨梅Morella rubra | MrLEC2 | KAB1221959.1 |
| 欧洲栓皮栎Quercus suber | QsLEC2 | XP_023881426.1 |
| 橡树Quercus lobata | QlLEC2 | XP_030973220.1 |
| 山嵛菜Eutrema salsugineum | EsLEC2 | XP_006415675.1 |
| 拟南芥琴亚种Arabidopsis lyrata subsp. | AlLEC2 | XP_020866568.1 |
| 拟南芥Arabidopsis thaliana | AtLEC2 | NP_564304.1 |
1.3 CsLEC2基因及编码蛋白的基本结构分析
用GSDS 2.0(http://gsds.cbi.pku.edu.cn/)分析CsLEC2基因外显子-内含子结构。运用Expasy数据库在线工具ProtParam(https://web.expasy. org/protparam/)分析CsLEC2编码蛋白的相对分子量和理论等电点等理化性质,以拟南芥AtLEC2作为对照,使用GeneDoc本地软件对CsLEC2蛋白进行多序列比对分析。应用在线网站TMHMM Server 2.0(http://www.cbs.dtu.dk/services/TMHMM/)、SignalP(http://www.cbs.dtu.dk/services/SignalP/)和Plant-mPLoc(http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/)对亚麻荠CsLEC2蛋白进行跨膜结构域、信号肽和亚细胞定位预测。利用在线网站SOPMA(https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopm.html)和SWISS-MODEL(https://swissmodel.expasy.org/)对亚麻荠CsLEC2蛋白进行二级结构和三级结构预测。
1.4 CsLEC2蛋白的保守结构域和系统进化分析
利用在线网站MEME Suite(http://meme-suite.org/tools/meme)对CsLEC2蛋白家族进行保守域预测,参数设置为默认参数;利用MEGA7.0软件对CsLEC2蛋白和其他物种LEC2蛋白进行多序列比对后,采用邻接法(neighbor-joining method)构建系统发育树。
1.5 CsLEC2启动子序列顺式元件分析
选取CsLEC2基因上游2 000bp的启动子序列,利用PlantCARE在线软件(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)对所选2 000bp范围内的启动子序列进行顺式元件预测。
1.6 CsLEC2基因的实时定量PCR表达分析
用TransZolPlant试剂盒(北京全式金生物技术有限公司)提取亚麻荠根、茎、叶、花及不同发育时期种子的RNA,用反转录试剂盒(ABM公司)将提取的RNA反转录为cDNA。用Premier 6.0软件设计CsLEC2基因特异性引物(表2),对CsLEC2基因进行RT-PCR,以亚麻荠CsActin作为内参基因。用试剂盒(Takara公司)进行qRT-PCR检测,20μL反应体系:定量上下引物各0.8μL、SYBR®Premix Ex TaqⅡ(2×) 10μL、ROX Reference Dye(50×)0.4μL、RNase-free H2O 7μL、cDNA模板1μL,每个样品设置3个生物学重复。荧光定量反应程序为95℃ 10min;95℃ 15s,60℃ 1min,40个循环;65℃ 5s,95℃ 15s。采用2-△△Cq方法计算每个基因的相对表达量,用SPSS 25.0软件分析显著性差异。
表2 CsLEC2基因的特异性引物
Table 2
| 基因名称Gene name | 引物(5′-3′) Primer (5′-3′) |
|---|---|
| CsActin | F: TTGGAAGGATCTGTACGGTAAC |
| R: TGTGAACGATTCCTGGACC | |
| CsLEC2.1 | F: CCTCTTCTAACGCAAACTCTGTCCA |
| R: GTTGACGAAATGAGTAGGCTACGAA | |
| CsLEC2.2 | F: ATGGATGCTAACAACAATCTCTCGC |
| R: TGTTTGGCCTTCACTCAAGACAAGA | |
| CsLEC2.3 | F: AGTGAACGAGAGGAACCA |
| R: CGGCTTGATAATGCTGATG |
1.7 CsLEC2下游基因预测
表3 预测基因在亚麻荠数据库中的编号
Table 3
| 基因名称Gene name | 亚麻荠数据库编号C. sativa database number |
|---|---|
| CsWRI1 | Csa06g028810.1 |
| CsFUS3 | Csa04g012470.1 |
| CsABI3-1 | Csa15g050420.1 |
| CsABI3-2 | Csa19g036630.1 |
| CsABI3-3 | Csa01g030760.1 |
| CsOLE1-1 | Csa12g028090.1 |
| CsOLE1-2 | Csa11g019460.1 |
| CsOLE2-1 | Csa11g057650.1 |
| CsOLE2-2 | Csa10g047190.1 |
| CsOLE2-3 | Csa09048s010.1 |
| CsOLE3-1 | Csa02g041750.1 |
| CsOLE3-2 | Csa18g022020.1 |
| CsOLE3-3 | Csa11g082710.1 |
2 结果与分析
2.1 CsLEC2家族基因结构与编码蛋白序列基本特征分析
表4 亚麻荠CsLEC2家族蛋白理化性质
Table 4
| 分析内容Analysis content | AtLEC2 | CsLEC2.1 | CsLEC2.2 | CsLEC2.3 |
|---|---|---|---|---|
| 数据库编号Database code | At1g28300.1 | Csa03g031590.1 | Csa14g035910.1 | Csa17g039140.1 |
| NCBI编号NCBI code | NP_564304.1 | XP_010499344.1 | XP_010460616.1 | XP_010478197.1 |
| 开放阅读框Open read frame (bp) | 1092 | 1092 | 1092 | 1044 |
| 染色体位置Location on chromosome | Chr1:9896566- 9900177 | Chr3:13175576- 13178955 | Chr14:14188382- 14191779 | Chr17:14245237- 14248478 |
| 蛋白长度The number of amino acids coding protein | 363 | 363 | 363 | 347 |
| 相对分子量Relative molecular weight (kDa) | 41.71 | 41.22 | 41.21 | 39.34 |
| 理论等电点Theoretical (pI) | 5.21 | 5.54 | 5.60 | 5.46 |
| 碱性氨基酸数Basic amino acid number | 36 | 34 | 35 | 36 |
| 酸性氨基酸数Acid amino acid number | 50 | 46 | 45 | 45 |
| 亲水性指数Hydropathy index | -0.818 | -0.556 | -0.654 | -0.600 |
| 氨基酸Amino acid (%) | Ser (10.2) | Ser (9.6) | Ser (9.6) | Ser (10.4) |
| Asn (9.1) | Asn (8.5) | Asn (8.5) | Asn (8.9) | |
| Leu (7.4) | Leu (8.5) | Leu (8.5) | Leu (8.4) |
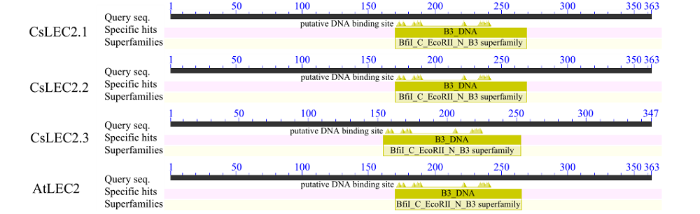
图1
图2
图2
亚麻荠CsLEC2家族蛋白功能结构域的分析
Fig.2
Analysis of functional domains of CsLEC2 family protein of C. sativa
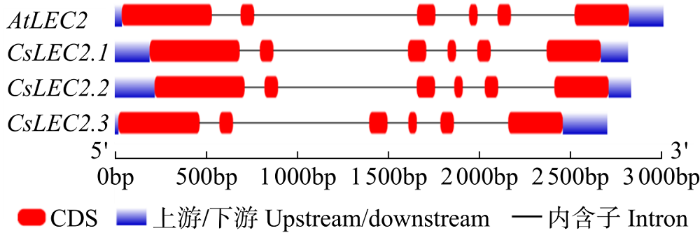
CsLEC2.1和CsLEC2.2开放阅读框均为1 092bp,编码363个氨基酸;CsLEC2.3开放阅读框为1 044bp,编码347个氨基酸。CsLEC2家族蛋白的相对分子量为39.34~41.71kDa,理论等电点为5.21~5.54(表4)。另外,3个CsLEC2蛋白的酸性氨基酸均多于碱性氨基酸,说明这3个CsLEC2蛋白在生理环境中均带负电。3个CsLEC2蛋白与拟南芥AtLEC2蛋白氨基酸含量最多的均为丝氨酸(Ser)、天冬酰胺(Asn)和亮氨酸(Leu),氨基酸的亲水性指数也一致。
用Softberry软件对CsLEC2蛋白进行亚细胞定位预测,结果显示3个CsLEC2蛋白和拟南芥AtLEC2蛋白主要定位在细胞核内。使用SignalP 4.1 Server和Tmhmm Server 2.0在线分析,显示亚麻荠CsLEC2和拟南芥AtLEC2蛋白均不含有跨膜区和对应的信号肽。
2.2 CsLEC2蛋白高级结构分析
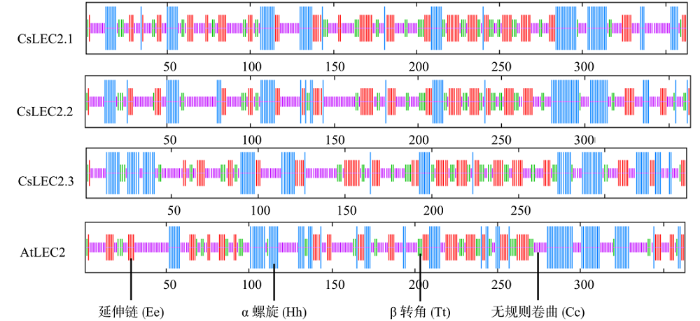
用在线工具SOPMA预测CsLEC2蛋白二级结构,发现亚麻荠3个CsLEC2蛋白的二级结构主要由无规则卷曲、α螺旋、延伸链和β-转角组成(图3)。亚麻荠CsLEC2.1蛋白二级结构由20.66%的α螺旋、13.77%的β转角、40.77%的无规则卷曲和24.79%延伸链组成;CsLEC2.2蛋白二级结构由20.94%的α螺旋、13.22%的β转角、43.80%的无规则卷曲和22.04%延伸链组成;CsLEC2.3蛋白二级结构由23.92%的α螺旋、14.12%的β转角、38.62%的无规则卷曲和23.34%延伸链组成。拟南芥AtLEC2蛋白二级结构由24.79%的α螺旋、11.57%的β转角、42.15%的无规则卷曲和21.49%延伸链组成。尽管亚麻荠CsLEC2和拟南芥AtLEC2蛋白二级结构中无规则卷曲占比例最大,β转角结构占比例最小,亚麻荠和拟南芥的LEC2蛋白在二级结构上的主体结构相似,但是在各个组成结构所占比例上存在一定差异。
图3
图3
亚麻荠CsLEC2蛋白二级结构
Fig.3
Comparison of secondary structure of CsLEC2 protein from C. sativa
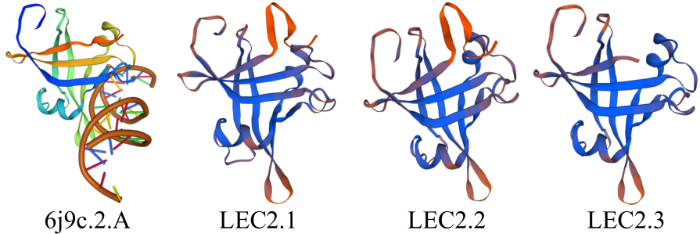
图4
图4
亚麻荠CsLEC2家族蛋白三级结构预测
Fig.4
Prediction of the tertiary structure of CSLEC2 family protein in C. sativa
2.3 CsLEC2氨基酸多序列比对
图5
图5
亚麻荠CsLEC2家族蛋白多序列比对
Fig.5
Multiple sequence alignment of CsLEC2 family proteins of C. sativa
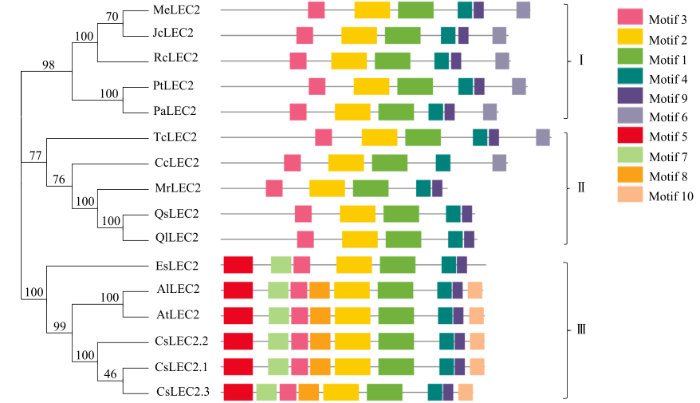
2.4 CsLEC2家族蛋白保守基序和系统进化分析
系统进化分析结果(图6)显示,各物种LEC2蛋白大致聚为3个亚族,亚麻荠CsLEC2蛋白与同属于十字花科的拟南芥AtLEC2蛋白、拟南芥琴亚种AlLEC2蛋白和山嵛菜EsLEC2蛋白均位于第III亚族中,且AtLEC2与AlLEC2蛋白的亲缘关系更近,可能在进化上有相同的进化来源。亚麻荠CsLEC2与其他两个亚族的麻风树JcLEC2、蓖麻RcLEC2、可可TcLEC2和克莱门柚CcLEC2的杂缘关系相距较远。
图6
图6
不同物种LEC2蛋白的系统进化树和保守基序
Fig.6
Phylogenetic tree and conserved motifs of LEC2 proteins from different species
在MEME网站预测到CsLEC2的保守结构域(表5)。亚麻荠CsLEC2蛋白、拟南芥AtLEC2蛋白和拟南芥琴亚种AlLEC2蛋白均含有除了motif 6的其他9个保守motif。山萮菜EsLEC2缺少motif 8和motif 10。第I亚族和第II亚族均不含第Ⅲ亚族蛋白的motif 5、motif 7、motif 8和motif 10。这4个motif可能是十字花科物种LEC2在进化上区别于其他物种的关键氨基酸残基位点。综上,MEME保守基序预测结果与进化树聚类结果完全一致,证明同属于十字花科的LEC2蛋白在进化上相对保守。
表5 保守基序信息
Table 5
| Motif编号Motif number | 氨基酸残基Amino acid residue |
|---|---|
| motif 1 | QSWSFKYKFWSNNKSRMYVLENTGEFVKQNGAEIGDFLTIYEDESKNLYF |
| motif 2 | YKFCTPDNKRLRVLLRKELKNSDVGSLGRIVLPKREAEGNLPTLSDKEGI |
| motif 3 | RRALDAYKTKVARCKRKLARQRSL |
| motif 4 | RDEEEASLALLIEQLRHKEQQ |
| motif 5 | FFPFSSSNANSVQEFAMDANNNLSHLTTMPTYDHHQAEPHH |
| motif 6 | YIDDCYSGLDVLPDVNRYNF |
| motif 7 | YSSDAYPQIPVSQTGSEFCSLVSNPNPCL |
| motif 8 | SSPNSSPDEVVDSKRQVMMLNMKNNVQIP |
| motif 9 | PNDLMGLTIDHQHHQ |
| motif 10 | SVDYAHVGSLDDQVSFDDIVW |
2.5 CsLEC2启动子的功能元件分析
选取CsLEC2基因上游2 000bp序列进行启动子分析,运用PlantCARE在线软件分析亚麻荠CsLEC2基因上游顺式元件(表6)。3个CsLEC2基因均有CAAT-box和TATA-box启动子基本元件,涉及光响应的顺式作用元件、厌氧诱导必需的调节元素、根特异性表达元件、雌激素应答元件和MYB结合元件等顺式元件,与激素响应相关的茉莉酸和生长素等顺式调节元件。
表6 CsLEC2基因上游启动子顺式元件
Table 6
| 顺式作用元件Cis-acting element | 核心序列Core sequence | 功能Function |
|---|---|---|
| CAAT-box | CAAT | 启动子和增强子区域常见的顺式作用元件 |
| TATA-box | TATA | 转录起始位点上游-30bp附近核心启动子元件 |
| ARE | AAACCA | 厌氧诱导必需的调节元素 |
| Box 4 | ATTAAT | 涉及光响应的保守DNA模块 |
| CGTCA-motif | CGTCA | MeJa响应的相关顺式作用调控元件 |
| TGACG-motif | TGACG | MeJa响应的相关顺式作用调控元件 |
| MBSI | aaaAaaC(G/C)GTTA | MYB结合位点,参与类黄酮生物合成基因的调控 |
| MYB | CAACCA | MYB结合元件 |
| TGA-element | AACGAC | 生长素响应元件 |
| ERE | ATTTCATA | 雌激素应答元件 |
| as-1 | TGACG | 根特异表达元件 |
2.6 CsLEC2基因的表达分析
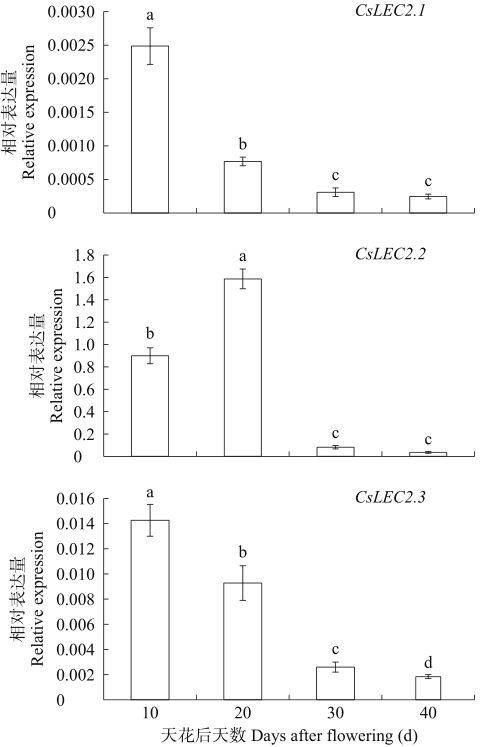
为进一步分析CsLEC2基因的时空表达特征,采用RT-PCR和qRT-PCR检测各基因在亚麻荠根、茎、叶、花及开花后不同发育时期的种子(10、20、30和40d)的表达量。结果如图7所示,内参基因(CsActin)在各组织中稳定高量表达,而CsLEC2.1、CsLEC2.2和CsLEC2.3基因均仅在开花后10和20d的发育种子中高量表达,表明这3个基因仅在亚麻荠发育中的种子中特异性表达。qRT-PCR结果(图8)表明,3个CsLEC2基因均在种子发育早期和早中期种子中高量表达,CsLEC2.2基因在开花后表达量逐渐升高,在开花后20d表达量最高,在开花后30和40d时表达量显著降低。CsLEC2.1和CsLEC2.3基因均在开花后10d表达量最高,表达量随着种子的逐渐成熟而显著降低,在种子发育的晚期(开花后40d左右)CsLEC2基因的表达量达到最低,其中CsLEC2.3基因的降低趋势较缓,在开花后20d的表达量明显高于CsLEC2.1基因。这3个基因总体表达趋势同半定量结果一致,推测这3个基因主要在种子发育早中期调控胚胎发育以及中后期种子油脂合成和积累等重要生理过程。
图7
图7
亚麻荠CsLEC2在不同组织和种子不同发育时期的RT-PCR结果
Fig.7
RT-PCR results of CsLEC2 expressions in various tissues and different developmental stages seeds of C. sativa
图8
图8
CsLEC2基因在种子不同发育时期的表达分析
不同小写字母表示在P<0.05水平上差异显著
Fig.8
Expression analysis of CsLEC2 at different seed developmental stages of C. sativa
Different lowercase letters indicate significantly difference at P < 0.05
2.7 CsLEC2调控的下游基因预测
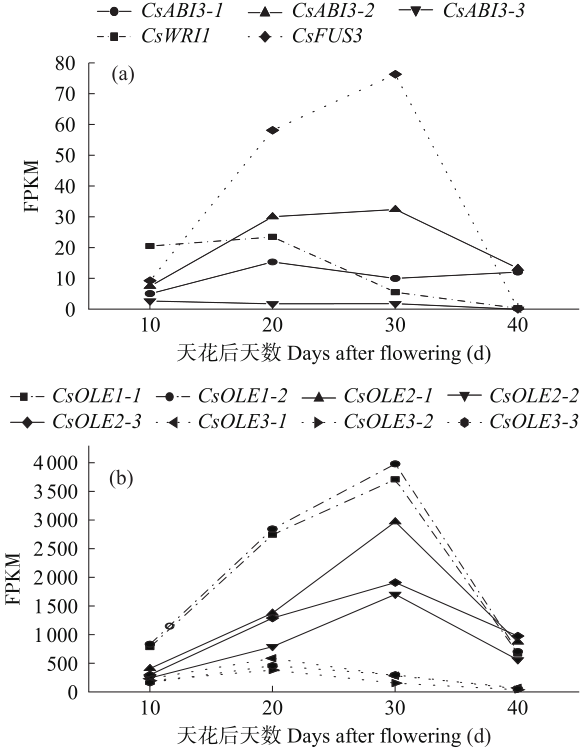
参与油脂生物合成基因的转录组数据显示,亚麻荠CsWRI1基因在各组织中均有转录表达,在种子中转录表达量最高,而CsFUS3、CsABI3和部分CsOLE基因均只在种子中转录表达。因此,CsFUS3、CsABI3和CsOLE基因与CsLEC2基因均是在种子中特异性表达的基因。根据亚麻荠转录组数据中CsWRI1、CsFUS3、CsABI3和CsOLE基因在种子不同发育时期的FPKM值,预测这些基因的表达模式(图9)。从图9(a)中可知,CsWRI1和CsFUS3-2基因在开花后10d转录表达值逐渐升高,在开花后20d转录表达值达到最高,在开花后30d到40d时转录表达值逐渐下降并趋于0,与亚麻荠CsLEC2.2的表达模式基本吻合。而2个CsFUS3基因与3个CsABI3基因在种子中的转录表达模式与3个CsLEC2基因的表达模式都不一致。图9(b)中3个CsOLE3基因的转录表达值变化趋势也与亚麻荠CsLEC2.2一致,而所有CsOLE1和CsOLE2基因的转录表达值趋势与3个CsLEC2基因的表达趋势均不一致。根据这几个基因与CsLEC2的共表达趋势,推测在亚麻荠种子发育时期CsLEC2.2转录因子可能直接调控下游CsWRI1和CsOLE3基因的表达。综上,CsLEC2基因在种子发育的早期可能也参与了对CsFUS3、CsABI3和亚麻荠中所有的CsOLE1和CsOLE2基因的调控,而种子发育后期表达趋势不完全一致的原因可能是在种子发育晚期增加了其他转录因子对这些基因的调控。
图9
图9
基因在亚麻荠种子不同发育时期的FPKM变化趋势
Fig.9
FPKM variation trend of genes at different seed developmental stages of C. sativa
3 讨论
3.1 CsLEC2的序列特征分析
目前对LEC2基因的功能研究多集中在拟南芥、玉米和甘蓝型油菜等植物上,而有关异源六倍体作物亚麻荠LEC2基因的研究较少。本研究在亚麻荠基因组数据库中共鉴定得到3个CsLEC2,与AtLEC2均属于B3超家族。AtLEC2含有B3结构域,是调控种子胚胎发育和储存营养物质的关键转录调控因子,能与RY-motif的DNA序列结合,而具有RY-motif的基因是种子特异性表达相关的基因[13]。LEC2基因可以正调控MYB115和MYB118的表达,而MYBs是胚乳中ω-7单不饱和脂肪酸合成的正向调节因子,在拟南芥lec2突变体种子中,油脂含量和蛋白质含量都较野生型有所下降[21]。基因结构分析显示,CsLEC2与AtLEC2基因结构相似。CsLEC2和AtLEC2理化性质也非常相似,均属于酸性、亲水性蛋白,且都不含有跨膜结构和信号肽。亚细胞定位分析表明,亚麻荠CsLEC2家族基因均定位在细胞核内,表明其在细胞核内起转录调控作用。三级结构表明CsLEC2蛋白与6j9c.2[20]模板蛋白(拟南芥转录因子LEC2-DNA复合物的晶体结构)相似度高,因此推测这3个CsLEC2蛋白与AtLEC2一样也具有调控种子胚胎发育和参与脂肪酸生物合成的功能。
Boulard等[22]研究表明,LEC2的B2结构域中存在保守残基TKxxARxxRxxAxxR,该保守残基可与LEC1中的D86K氨基酸残基结合形成异聚体,参与基因表达调控。多序列比对结果显示,除B3保守结构域外,亚麻荠CsLEC2家族蛋白也都存在B2结构域的保守残基TKxxARxxRxxAxxR,证明CsLEC2也可能与CsLEC1相互配合参与对下游基因的调控。
系统进化树结果表明,亚麻荠CsLEC2蛋白与同属于十字花科的拟南芥AtLEC2蛋白、拟南芥琴亚种AlLEC2蛋白及山嵛菜EsLEC2的亲缘关系较近,推测CsLEC2蛋白与AtLEC2蛋白可能参与调节油脂的积累及控制脂肪酸生物合成[6]。MEME预测的结果与进化树聚类的结果一致,均表明亚麻荠CsLEC2家族蛋白的保守性较高。
亚麻荠CsLEC2基因上游启动子元件分析表明,除含有启动子的基本元件外,还有涉及光响应的顺式作用元件、厌氧诱导必需的调节元素、根特异性表达元件、雌激素应答元件、MYB结合元件等顺式元件和与激素响应相关的茉莉酸和生长素等顺式调节元件。许多MYB蛋白和含有MYB结合元件的基因对拟南芥、玉米、棉花、苹果和胡杨都有干旱反应[23]。杨英军等[24]对proteinase omega基因5′上游调控序列分析,发现多种逆境应答调控元件ERE、ABRE和HSE等。果天宇等[25]发现ZmCOL3pro217启动子中含有分生组织特异性、光响应和胁迫响应等顺式作用元件,推测其参与玉米开花调控。过表达LEC2基因可激活一些生长素响应基因的表达[16]。亚麻荠CsLEC2启动子序列的顺式作用元件分析表明,CsLEC2可能参与逆境环境胁迫应答,同时也可以通过光响应参与亚麻荠开花调控。
3.2 CsLEC2的表达模式分析
本研究发现,3个亚麻荠CsLEC2基因都只在种子中表达。实时荧光定量结果显示,在亚麻荠不同发育时期的种子中CsLEC2的表达均有差异,但都是在未成熟的种子中高表达,随着种子的发育成熟表达量逐渐降低甚至不表达。大豆GmLEC2也是在未成熟的种子中表达量最高[26],与亚麻荠CsLEC2表达模式相同。因此推测亚麻荠CsLEC2基因与种子中油脂的积累相关,并且是在亚麻荠种子发育的早中期通过调节相关基因的表达而发挥作用。
3.3 CsLEC2的下游基因预测分析
通过对几个基因不同发育时期的转录表达值变化趋势分析,本研究发现亚麻荠CsWRI1和CsOLE3基因在转录组数据中的转录表达值变化趋势与亚麻荠CsLEC2.2相似。Mendoza等[13]过表达拟南芥AtLEC2提高了拟南芥中LEC1、FUS3和ABI3基因的表达量。Kim等[14]在拟南芥中超表达蓖麻RcLEC2基因,激活了油体蛋白合成相关基因的表达,种子含油量增加了约2.7倍。Baud等[15]在拟南芥中证实了AtLEC2能够通过调控下游AtWRI1基因的表达来调控脂肪酸合成相关基因的转录表达。这些都表明LEC2转录因子可以调控下游基因的表达。由本研究结果推测,在亚麻荠种子发育时期CsWRI和CsOLE3基因上游的转录调控因子为CsLEC2.2。而CsWRI1基因在亚麻荠根、茎、叶和花组织中也有表达,表明在这些组织中可能存在其他转录调控因子。
4 结论
以具有调控种子发育过程和油脂产量功能的拟南芥AtLEC2蛋白序列为参考序列,鉴定得到了3个亚麻荠CsLEC2基因,通过生物信息学分析发现亚麻荠CsLEC2蛋白与拟南芥AtLEC2蛋白有着相似的功能,通过RT-PCR和qRT-PCR表达特性分析发现亚麻荠CsLEC2在种子中特异表达,并且在种子发育的早期和早中期高表达。对几种不同基因在转录组数据的转录表达值的变化趋势分析发现亚麻荠CsLEC2.2转录因子可能直接调控下游CsWRI1和CsOLE3基因的表达。
参考文献
Identification of three genes encoding microsomal oleate desaturases (FAD2) from the oil seed crop Camelina sativa
DOI:10.1016/j.plaphy.2010.12.004 URL [本文引用: 1]
Generation of transgenic plants of a potential oil seed crop Camelina sativa by Agrobacterium-mediated transformation
DOI:10.1007/s00299-007-0454-0
URL
[本文引用: 1]

Camelina sativa is an alternative oilseed crop that can be used as a potential low-cost biofuel crop or a source of health promoting omega-3 fatty acids. Currently, the fatty acid composition of camelina does not uniquely fit any particular uses, thus limit its commercial value and large-scale production. In order to improve oil quality and other agronomic characters, we have developed an efficient and simple in planta method to generate transgenic camelina plants. The method included Agrobacterium-mediated inoculation of plants at early flowering stage along with a vacuum infiltration procedure. We used a fluorescent protein (DsRed) as a visual selection marker, which allowed us to conveniently screen mature transgenic seeds from a large number of untransformed seeds. Using this method, over 1% of transgenic seeds can be obtained. Genetic analysis revealed that most of transgenic plants contain a single copy of transgene. In addition, we also demonstrated that transgenic camelina seeds produced novel hydroxy fatty acids by transforming a castor fatty acid hydroxylase. In conclusion, our results provide a rapid means to genetically improve agronomic characters of camelina, including fatty acid profiles of its seed oils. Camelina may serve as a potential industrial crop to produce novel biotechnology products.]]>
Genome-wide analysis of the ERF gene family in Arabidopsis and rice
DOI:10.1104/pp.105.073783
URL
PMID:16407444
[本文引用: 1]

Genes in the ERF family encode transcriptional regulators with a variety of functions involved in the developmental and physiological processes in plants. In this study, a comprehensive computational analysis identified 122 and 139 ERF family genes in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa L. subsp. japonica), respectively. A complete overview of this gene family in Arabidopsis is presented, including the gene structures, phylogeny, chromosome locations, and conserved motifs. In addition, a comparative analysis between these genes in Arabidopsis and rice was performed. As a result of these analyses, the ERF families in Arabidopsis and rice were divided into 12 and 15 groups, respectively, and several of these groups were further divided into subgroups. Based on the observation that 11 of these groups were present in both Arabidopsis and rice, it was concluded that the major functional diversification within the ERF family predated the monocot/dicot divergence. In contrast, some groups/subgroups are species specific. We discuss the relationship between the structure and function of the ERF family proteins based on these results and published information. It was further concluded that the expansion of the ERF family in plants might have been due to chromosomal/segmental duplication and tandem duplication, as well as more ancient transposition and homing. These results will be useful for future functional analyses of the ERF family genes.
A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons
DOI:10.1126/science.258.5088.1647
URL
PMID:17742538
[本文引用: 1]

Cotyledons are specialized leaves produced during plant embryogenesis. Cotyledons and leaves typically differ in morphology, ultrastructure, and patterns of gene expression. The leafy cotyledon (Iec) mutant of Arabidopsis thaliana fails to maintain this distinction between embryonic and vegetative patterns of plant development. Mutant embryos are phenotypically abnormal, occasionally viviparous, and intolerant of desiccation. Mutant cotyledons produce trichomes characteristic of leaves, lack embryo-specific protein bodies, and exhibit a vascular pattern intermediate between that of leaves and cotyledons. These results suggest that lec cotyledons are partially transformed into leaves and that the wild-type gene (LEC) functions to activate a wide range of embryo-specific pathways in higher plants.
LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development
LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis,via YUCCA-mediated auxin biosynthesis
DOI:10.1007/s00425-013-1892-2
URL
[本文引用: 1]

The LEAFY COTYLEDON2 (LEC2) transcription factor with a plant-specific B3 domain plays a central role in zygotic and somatic embryogenesis (SE). LEC2 overexpression induced in planta leads to spontaneous somatic embryo formation, but impairs the embryogenic response of explants cultured in vitro under auxin treatment. The auxin-related functions of LEC2 appear during SE induction, and the aim of the present study was to gain further insights into this phenomenon. To this end, the effect of LEC2 overexpression on the morphogenic responses of Arabidopsis explants cultured in vitro under different auxin treatments was evaluated. The expression profiles of the auxin biosynthesis genes were analysed in embryogenic cultures with respect to LEC2 activity. The results showed that LEC2 overexpression severely modifies the requirement of cultured explants for an exogenous auxin concentration at a level that is effective in SE induction and suggested an increase in the auxin content in 35S::LEC2-GR transgenic explants. The assumption of an LEC2 promoted increase in endogenous auxin in cultured explants was further supported by the expression profiling of the genes involved in auxin biosynthesis. The analysis indicated that YUCCAs and TAA1, working in the IPA-YUC auxin biosynthesis pathway, are associated with SE induction, and that the expression of three YUCCA genes (YUC1, YUC4 and YUC10) is associated with LEC2 activity. The results also suggest that the IAOx-mediated auxin biosynthesis pathway involving ATR1/MYB34 and CYP79B2 does not seem to be involved in SE induction. We conclude that de novo auxin production via the tryptophan-dependent IPA-YUC auxin biosynthesis pathway is implicated in SE induction, and that LEC2 plays a key role in this mechanism.
The plant B3 superfamily
DOI:10.1016/j.tplants.2008.09.006
URL
[本文引用: 1]

Arabidopsis, there are 118 B3 genes, and in rice there are 91 B3 genes. The B3 domain is present in genes from gymnosperms, mosses and green algae, indicating that the B3 domain evolved on the plant lineage before multicellularity. The aim of this review is to phylogenetically characterize the members of the B3 family in Arabidopsis and rice and to review the function of the B3 genes that have been studied to date.]]>
Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by L-AFL proteins in Arabidopsis seed
DOI:10.1104/pp.16.00034
URL
PMID:27208266
[本文引用: 1]

In Arabidopsis (Arabidopsis thaliana), transcriptional control of seed maturation involves three related regulators with a B3 domain, namely LEAFY COTYLEDON2 (LEC2), ABSCISIC ACID INSENSITIVE3 (ABI3), and FUSCA3 (ABI3/FUS3/LEC2 [AFLs]). Although genetic analyses have demonstrated partially overlapping functions of these regulators, the underlying molecular mechanisms remained elusive. The results presented here confirmed that the three proteins bind RY DNA elements (with a 5'-CATG-3' core sequence) but with different specificities for flanking nucleotides. In planta as in the moss Physcomitrella patens protoplasts, the presence of RY-like (RYL) elements is necessary but not sufficient for the regulation of the OLEOSIN1 (OLE1) promoter by the B3 AFLs. G box-like domains, located in the vicinity of the RYL elements, also are required for proper activation of the promoter, suggesting that several proteins are involved. Consistent with this idea, LEC2 and ABI3 showed synergistic effects on the activation of the OLE1 promoter. What is more, LEC1 (a homolog of the NF-YB subunit of the CCAAT-binding complex) further enhanced the activation of this target promoter in the presence of LEC2 and ABI3. Finally, recombinant LEC1 and LEC2 proteins produced in Arabidopsis protoplasts could form a ternary complex with NF-YC2 in vitro, providing a molecular explanation for their functional interactions. Taken together, these results allow us to propose a molecular model for the transcriptional regulation of seed genes by the L-AFL proteins, based on the formation of regulatory multiprotein complexes between NF-YBs, which carry a specific aspartate-55 residue, and B3 transcription factors.
植物中油脂调节基因ABSCISIC ACID INSENSITIVE3、FUSCA3和LEAFY COTYLEDON2的鉴定、系统进化和结构特征分析
LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves
DOI:10.1016/j.febslet.2005.07.037
URL
PMID:16107256
[本文引用: 3]

LEAFY COTYLEDON 2 (LEC2) is a key regulator of seed maturation in Arabidopsis. To unravel some of its complex pleiotropic functions, analyses were performed with transgenic plants expressing an inducible LEC2:GR protein. The chimeric protein is functional and can complement lec2 mutation. Interestingly, the induction of LEC2 leads to the accumulation of storage oil in leaves. In addition, short-term induction and use of translation inhibitors allowed to demonstrate that LEC2 can directly trigger the accumulation of seed specific mRNAs. Consistent with these results, the expression of three other major seed regulators namely, LEC1, FUS3, and ABI3 were also induced by LEC2 activation.
Ectopic overexpression of castor bean LEAFY COTYLEDON2 (LEC2) in Arabidopsis triggers the expression of genes that encode regulators of seed maturation and oil body proteins in vegetative tissues
DOI:10.1016/j.fob.2013.11.003
URL
PMID:24363987
[本文引用: 2]

The LEAFY COTYLEDON2 (LEC2) gene plays critically important regulatory roles during both early and late embryonic development. Here, we report the identification of the LEC2 gene from the castor bean plant (Ricinus communis), and characterize the effects of its overexpression on gene regulation and lipid metabolism in transgenic Arabidopsis plants. LEC2 exists as a single-copy gene in castor bean, is expressed predominantly in embryos, and encodes a protein with a conserved B3 domain, but different N- and C-terminal domains to those found in LEC2 from Arabidopsis. Ectopic overexpression of LEC2 from castor bean under the control of the cauliflower mosaic virus (CaMV) 35S promoter in Arabidopsis plants induces the accumulation of transcripts that encodes five major transcription factors (the LEAFY COTYLEDON1 (LEC1), LEAFY COTYLEDON1-LIKE (L1L), FUSCA3 (FUS3), and ABSCISIC ACID INSENSITIVE 3 (ABI3) transcripts for seed maturation, and WRINKELED1 (WRI1) transcripts for fatty acid biosynthesis), as well as OLEOSIN transcripts for the formation of oil bodies in vegetative tissues. Transgenic Arabidopsis plants that express the LEC2 gene from castor bean show a range of dose-dependent morphological phenotypes and effects on the expression of LEC2-regulated genes during seedling establishment and vegetative growth. Expression of castor bean LEC2 in Arabidopsis increased the expression of fatty acid elongase 1 (FAE1) and induced the accumulation of triacylglycerols, especially those containing the seed-specific fatty acid, eicosenoic acid (20:1(Delta11)), in vegetative tissues.
WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis
DOI:10.1111/j.1365-313X.2007.03092.x
URL
PMID:17419836
[本文引用: 2]

The WRINKLED1 (WRI1) transcription factor has been shown to play a role of the utmost importance during oil accumulation in maturing seeds of Arabidopsis thaliana. However, little is known about the regulatory processes involved. In this paper, comprehensive functional analyses of three new mutants corresponding to null alleles of wri1 confirm that the induction of WRI1 is a prerequisite for fatty acid synthesis and is important for normal embryo development. The strong expression of WRI1 specifically detected at the onset of the maturation phase in oil-accumulating tissues of A. thaliana seeds is fully consistent with this function. Complementation experiments carried out with various seed-specific promoters emphasized the importance of a tight regulation of WRI1 expression for proper oil accumulation, raising the question of the factors controlling WRI1 transcription. Interestingly, molecular and genetic analyses using an inducible system demonstrated that WRI1 is a target of LEAFY COTYLEDON2 and is necessary for the regulatory action of LEC2 towards fatty acid metabolism. In addition to this, quantitative RT-PCR experiments suggested that several genes encoding enzymes of late glycolysis, the fatty acid synthesis pathway, and the biotin and lipoic acid biosynthetic pathways are targets of WRI1. Taken together, these results indicate new relationships in the regulatory model for the control of oil synthesis in maturing A. thaliana seeds. In addition, they exemplify how metabolic and developmental processes affecting the developing embryo can be coordinated at the molecular level.
Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis
Regulation of storage protein gene expression in Arabidopsis
Mutation of the transcription factor LEAFY COTYLEDON 2 alters the chemical composition of Arabidopsis seeds,decreasing oil and protein content,while maintaining high levels of starch and sucrose in mature seeds
DOI:10.1016/j.jplph.2011.05.003
URL
[本文引用: 1]

The transcription factor LEAFY COTYLEDON 2 (LEC2: At1g28300) is preferentially expressed in developing seeds of Arabidopsis. Detailed biochemical analysis of a loss-of-function lec2 mutant was carried out in seeds 6-21 days after flowering (OAF). In comparison to wild type controls, lec2 seeds had 15% less protein and 30% less oil, but accumulated 140% more sucrose and >5-fold more starch. We also quantified biomass and carbohydrates in the seed coat and embryo. The lec2 mutant had smaller seeds and an altered proportion of dry weight (bigger seed coat and smaller embryos). Mutant plants produced less mature seeds per silique and the harvest index was reduced. Soluble sugars (glucose, fructose and sucrose) was accumulated in the seed coat of the lec2 mutant, whereas the opposite effect was observed in the embryos (decrease in comparison to wild type). The rate of starch synthesis increased during early development, whereas the rate of starch degradation was diminished during late development, leading to higher residual starch in mature seed of the mutant. Starch accumulated in both seed coat and embryo. Homozygous mutant plants produced seeds that could germinate well if they were harvested immaturely, whereas seeds that became dry during maturity lost their germination efficiency very rapidly. We conclude that the LEC2 transcription factor not only controls cotyledon identity and morphology as previously reported, but also alters: (1) the delivery of photosynthates from the seed coat to the embryo (sink strength), (2) carbon partitioning towards different storage compounds (oil, proteins and carbohydrates), (3) the rate of starch synthesis and degradation in developing seeds and (4) germination capacity of dry seeds. (C) 2011 Elsevier GmbH.
The developmental transcriptome atlas of the biofuel crop Camelina Sativa
DOI:10.1111/tpj.13302
URL
PMID:27513981
[本文引用: 1]

Camelina sativa is currently being embraced as a viable industrial bio-platform crop due to a number of desirable agronomic attributes and the unique fatty acid profile of the seed oil that has applications for food, feed and biofuel. The recent completion of the reference genome sequence of C. sativa identified a young hexaploid genome. To complement this work, we have generated a genome-wide developmental transcriptome map by RNA sequencing of 12 different tissues covering major developmental stages during the life cycle of C. sativa. We have generated a digital atlas of this comprehensive transcriptome resource that enables interactive visualization of expression data through a searchable database of electronic fluorescent pictographs (eFP browser). An analysis of this dataset supported expression of 88% of the annotated genes in C. sativa and provided a global overview of the complex architecture of temporal and spatial gene expression patterns active during development. Conventional differential gene expression analysis combined with weighted gene expression network analysis uncovered similarities as well as differences in gene expression patterns between different tissues and identified tissue-specific genes and network modules. A high-quality census of transcription factors, analysis of alternative splicing and tissue-specific genome dominance provided insight into the transcriptional dynamics and sub-genome interplay among the well-preserved triplicated repertoire of homeologous loci. The comprehensive transcriptome atlas in combination with the reference genome sequence provides a powerful resource for genomics research which can be leveraged to identify functional associations between genes and understand the regulatory networks underlying developmental processes.
Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis
DOI:10.1038/s41477-019-0402-3
URL
PMID:30962525
[本文引用: 2]

Some overwintering plants acquire competence to flower, after experiencing prolonged cold in winter, through a process termed vernalization. In the crucifer plant Arabidopsis thaliana, prolonged cold induces chromatin-mediated silencing of the potent floral repressor FLOWERING LOCUS C (FLC) by Polycomb proteins. This vernalized state is epigenetically maintained or 'memorized' in warm rendering plants competent to flower in spring, but is reset in the next generation. Here, we show that in early embryogenesis, two homologous B3 domain transcription factors LEAFY COTYLEDON 2 (LEC2) and FUSCA3 (FUS3) compete against two repressive B3-containing epigenome readers and Polycomb partners known as VAL1 and VAL2 for the cis-regulatory cold memory element (CME) of FLC to disrupt Polycomb silencing. Consistently, crystal structures of B3-CME complexes show that B3FUS3, B3LEC2 and B3VAL1 employ a nearly identical binding interface for CME. We further found that LEC2 and FUS3 recruit the scaffold protein FRIGIDA in association with active chromatin modifiers to establish an active chromatin state at FLC, which results in resetting of the silenced FLC to active and erasing the epigenetic parental memory of winter cold in early embryos. Following embryo development, LEC2 and FUS3 are developmentally silenced throughout post-embryonic stages, enabling VALs to bind to the CME again at seedling stages at which plants experience winter cold. Our findings illustrate how overwintering crucifer annuals or biennials in temperate climates employ a subfamily of B3 domain proteins to switch on, off and on again the expression of a key flowering gene in the embryo-to-plant-to-embryo cycle, and thus to synchronize growth and development with seasonal temperature changes in their life cycles.
Transcriptional activation of two delta-9 palmitoyl-ACP desaturase genes by MYB115 and MYB118 is critical for biosynthesis of omega-7 monounsaturated fatty acids in the endosperm of Arabidopsis seeds
DOI:10.1105/tpc.16.00612
URL
PMID:27681170
[本文引用: 2]

In angiosperms, double fertilization of the embryo sac initiates the development of the embryo and the endosperm. In Arabidopsis thaliana, an exalbuminous species, the endosperm is reduced to one cell layer during seed maturation and reserves such as oil are massively deposited in the enlarging embryo. Here, we consider the strikingly different fatty acid (FA) compositions of the oils stored in the two zygotic tissues. Endosperm oil is enriched in omega-7 monounsaturated FAs, that represent more than 20 mol% of total FAs, whereas these molecular species are 10-fold less abundant in the embryo. Two closely related transcription factors, MYB118 and MYB115, are transcriptionally induced at the onset of the maturation phase in the endosperm and share a set of transcriptional targets. Interestingly, the endosperm oil of myb115 myb118 double mutants lacks omega-7 FAs. The identification of two Delta9 palmitoyl-ACP desaturases responsible for omega-7 FA biosynthesis, which are activated by MYB115 and MYB118 in the endosperm, allows us to propose a model for the transcriptional control of oil FA composition in this tissue. In addition, an initial characterization of the structure-function relationship for these desaturases reveals that their particular substrate specificity is conferred by amino acid residues lining their substrate pocket that distinguish them from the archetype Delta9 stearoyl-ACP desaturase.
LEC1 (NF-YB9) directly interacts with LEC2 to control gene expression in seed
DOI:10.1016/j.bbagrm.2018.03.005
URL
PMID:29580949
[本文引用: 1]

The LAFL transcription factors LEC2, ABI3, FUS3 and LEC1 are master regulators of seed development. LEC2, ABI3 and FUS3 are closely related proteins that contain a B3-type DNA binding domain. We have previously shown that LEC1 (a NF-YB type protein) can increase LEC2 and ABI3 but not FUS3 activity. Interestingly, FUS3, LEC2 and ABI3 contain a B2 domain, the function of which remains elusive. We showed that LEC1 and LEC2 partially co-localised in the nucleus of developing embryos. By comparing protein sequences from various species, we identified within the B2 domains a set of highly conserved residues (i.e. TKxxARxxRxxAxxR). This domain directly interacts with LEC1 in yeast. Mutations of the conserved amino acids of the motif in the B2 domain abolished this interaction both in yeast and in moss protoplasts and did not alter the nuclear localisation of LEC2 in planta. Conversely, the mutations of key amino acids for the function of LEC1 in planta (D86K) prevented the interaction with LEC2. These results provide molecular evidences for the binding of LEC1 to B2-domain containing transcription factors, to form heteromers, involved in the control of gene expression.
Comprehensive transcriptomic study on horse gram (Macrotyloma uniflorum):de novo assembly,functional characterization and comparative analysis in relation to drought stress
DOI:10.1186/1471-2164-14-647
URL
PMID:24059455
[本文引用: 1]

BACKGROUND: Drought tolerance is an attribute maintained in plants by cross-talk between multiple and cascading metabolic pathways. Without a sequenced genome available for horse gram, it is difficult to comprehend such complex networks and intercalated genes associated with drought tolerance of horse gram (Macrotyloma uniflorum). Therefore, de novo transcriptome discovery and associated analyses was done for this highly drought tolerant yet under exploited legume to decipher its genetic makeup. RESULTS: Eight samples comprising of shoot and root tissues of two horse gram genotypes (drought-sensitive; M-191 and drought-tolerant; M-249) were used for comparison under control and polyethylene glycol-induced drought stress conditions. Using Illumina sequencing technology, a total of 229,297,896 paired end read pairs were generated and utilized for de novo assembly of horse gram. Significant BLAST hits were obtained for 26,045 transcripts while, 3,558 transcripts had no hits but contained important conserved domains. A total of 21,887 unigenes were identified. SSRs containing sequences covered 16.25% of the transcriptome with predominant tri- and mono-nucleotides (43%). The total GC content of the transcriptome was found to be 43.44%. Under Gene Ontology response to stimulus, DNA binding and catalytic activity was highly expressed during drought stress conditions. Serine/threonine protein kinase was found to dominate in Enzyme Classification while pathways belonging to ribosome metabolism followed by plant pathogen interaction and plant hormone signal transduction were predominant in Kyoto Encyclopedia of Genes and Genomes analysis. Independent search on plant metabolic network pathways suggested valine degradation, gluconeogenesis and purine nucleotide degradation to be highly influenced under drought stress in horse gram. Transcription factors belonging to NAC, MYB-related, and WRKY families were found highly represented under drought stress. qRT-PCR validated the expression profile for 9 out of 10 genes analyzed in response to drought stress. CONCLUSIONS: De novo transcriptome discovery and analysis has generated enormous information over horse gram genomics. The genes and pathways identified suggest efficient regulation leading to active adaptation as a basal defense response against drought stress by horse gram. The knowledge generated can be further utilized for exploring other underexploited plants for stress responsive genes and improving plant tolerance.