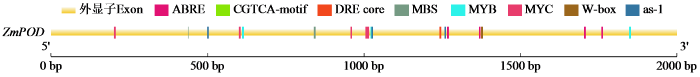作物对干旱胁迫的响应是由多基因控制的复杂数量性状。干旱对玉米生长的影响深刻且广泛,不仅会改变玉米表型,还会影响其生理生化过程。在干旱胁迫条件下,玉米在生理生化层面会发生一系列变化,以减轻自身受伤害的程度,例如增强保护酶活性、降低光合作用效率以及增加渗透调节物质含量等[8]。过氧化物酶(POD)是一类分布广泛的酶,可通过催化过氧化氢(H2O2)与各种有机、无机化合物间的氧化还原反应,在活性氧(ROS)的生成与清除过程中发挥重要作用[9]。根据其蛋白质结构,POD可分为2类,包括血红素过氧化物酶和非血红素过氧化物酶[10]。近年来,随着分子生物学研究的持续深入,玉米和水稻等多个物种测序的完成,以及生物信息学的飞速发展,人们对POD的结构、生理生化活性、表达调控机制及生物学功能有了更为深入的认识。
POD根据其序列和催化性质可进一步分为3类,这3类POD同属1个超家族,均含有由原卟啉IX和Fe(III)结合形成的血红素辅基,且三维结构高度相似[11
随着全球温室效应的加剧,可利用水资源日益减少,干旱已成为限制玉米生长发育及产量提升的重要非生物胁迫因子。玉米抗旱性属于数量性状,针对其开展遗传分析、基因定位和功能解析具有复杂性和艰巨性。目前,玉米生产中可利用的抗旱基因资源较少,因此,玉米抗旱基因的分离与功能解析已成为抗旱育种工作亟待突破的核心问题和技术瓶颈。本课题组前期以玉米耐旱品种农单476为材料,通过对比干旱处理与正常条件下的转录组和蛋白组数据,成功挖掘到1个受干旱调控的过氧化物酶基因ZmPOD。基于此,本研究进一步克隆ZmPOD基因,并开展生物信息学分析,通过拟南芥过表达体系进行功能验证。试验结果初步阐明了ZmPOD基因在干旱胁迫应答中的作用,为深入解析POD的生物学功能提供理论依据。
1 材料与方法
1.1 ZmPOD基因的克隆
种植农单476的亲本农系3435,使用全能型植物RNA提取试剂盒OminiPlant RNA Kit(CW2598S,北京康为世纪生物科技有限公司)提取叶片总RNA,使用反转录试剂盒PrimeScriptTM RT-PCR Kit[RR014A,宝生物工程(大连)有限公司]构建测序所需的cDNA文库。根据ZmPOD基因的CDS序列,利用Primer Premier 5软件设计克隆基因的引物。ZmPOD-F引物序列(5′-3′)为ATGGCTAAGGAAAGCAAGCTA;ZmPOD-R引物序列(5'-3')为CTAGCTCGCGGCGTAGCC。使用PrimeSTAR® Max DNA Polymerase试剂盒[R045Q,宝生物工程(大连)有限公司]对目的基因进行PCR扩增,扩增体系为50 µL,包含cDNA模板2 µL、特异性引物F(10 μmol/L)2 µL、特异性引物R(10 μmol/L)2 µL、PrimeSTAR Max Premix 25 µL和ddH2O 19 µL。扩增程序为:95 ℃预变性5 min;随后进行95 ℃ 15 s、56 ℃ 15 s、68 ℃ 30 s的循环,共计35次;最后68 ℃延伸10 min,并在12 ℃条件保存。对扩增的目的片段进行1%琼脂糖凝胶电泳检测,观察目的片段的大小及位置,使用M5 HiPure Gel Extraction Kit试剂盒(MF029-01,北京聚合美生物科技有限公司)进行胶回收与纯化,将纯化后的产物送至生工生物工程(上海)股份有限公司进行测序。
1.2 ZmPOD蛋白序列及结构分析
利用ProtParam网站对ZmPOD的分子量、氨基酸组成和等电点等理化性质进行分析;分别利用SOPMA和SWISS-MODEL在线网站预测ZmPOD蛋白的二级和三级结构;利用TMHMM在线网站进行ZmPOD蛋白序列跨膜区域预测;利用SignalP 5.0在线网站预测ZmPOD蛋白信号肽;利用ProtScale网站预测ZmPOD蛋白亲水性/疏水性;利用NetPhos 3.1 Server网站统计丝氨酸、苏氨酸和酪氨酸的磷酸化位点。
1.3 ZmPOD蛋白序列比对
利用NCBI数据库查询ZmPOD基因的编码氨基酸序列,利用NCBI在线软件Protein BLAST比对与ZmPOD基因编码氨基酸序列同源性较高的前20个物种,通过DNAMAN软件进行多重比较。
1.4 ZmPOD基因的启动子克隆与分析
利用NCBI数据库查询ZmPOD基因起始密码子上游2000 bp的序列,利用Primer Premier 5软件设计克隆启动子的引物。ZmPOD-F引物序列(5'-3')为CACCACAATCCATGCGAGTACAAG;ZmPOD- R引物序列(5'-3')为CAGTAGCAAGAGAGCTTC CGATATC。使用PrimeSTAR® Max DNA Polymerase试剂盒[R045Q,宝生物工程(大连)有限公司]对目的基因进行PCR扩增。以农系3435的DNA为模板,扩增体系为50 µL,包含cDNA模板2 µL、特异性引物F(10 μmol/L)2 µL、特异性引物R(10 μmol/L)2 µL、PrimerSTAR Max Premix 25 µL和ddH2O 19 µL。扩增程序为:95 ℃预变性5 min;随后进行95 ℃ 15 s、62 ℃ 15 s、72 ℃ 30 s的循环,共计35次;最后72 ℃延伸10 min,并在12 ℃条件保存。对扩增的目的片段进行1%琼脂糖凝胶电泳检测,观察目的片段的大小及位置,使用M5 HiPure Gel Extraction Kit试剂盒(MF029-01,北京聚合美生物科技有限公司)进行胶回收与纯化。将纯化后的产物送至生工生物工程(上海)股份有限公司进行测序。
使用Plant-CARE在线网站预测ZmPOD基因启动子区的顺式作用元件,并利用GSDS 2.0在线网站将启动子区的顺式作用元件绘制成图。
1.5 ZmPOD拟南芥过表达材料的获得
使用无缝克隆试剂盒Minerva Super Fusion Cloning Kit(M2026L,上海泽叶生物科技有限公司)将目的片段与pGreen-35s-6HA载体进行连接。连接体系为Super Fusion Cloning Mix 5 µL,目的片段50~200 ng,并用ddH2O补足至10 µL。将连接体系转化大肠杆菌菌株Top10以获得重组质粒,验证后将其转化农杆菌GV3101(pSoup),最后使用农杆菌介导的蘸花法获得拟南芥阳性苗。待拟南芥成熟,收获种子并干燥处理,所获种子即为T0代。
将收获的T0代拟南芥种子于4 ℃冰箱春化2~4 d,之后将其高密度、均匀地种植于营养土和蛭石比例为1:1的混合物中。待幼苗长出2片发育良好的真叶时,喷施Basta除草剂(25 mg/L草铵膦,工作浓度为1:2000),每隔1 d喷施1次,共3次,筛选转基因阳性植株。随后,对筛选到的阳性植株进行常规培养一段时间后,提取叶片中的DNA(CTAB法)和RNA,分别用于PCR和qRT-PCR鉴定。按上述方法繁育种子,获得T3代转基因植株,用于后续的表型测定。
1.6 拟南芥表型鉴定
为验证ZmPOD基因的功能,将野生型拟南芥(WT)及3个转基因株系(L1、L11、L12)的种子经春化处理后,用5% NaClO消毒5 min,无菌水冲洗2~3次,将其置于1/2 MS固体培养基中培养7 d。取部分幼苗移至0、200和300 mmol/L甘露醇的1/2 MS培养基中,竖直培养7 d,观察拟南芥根系生长状态并测量根长。
将另一部分幼苗移栽至土壤和水分条件一致的穴盘中,对筛选到的阳性苗正常培养一段时间后,提取叶片RNA。使用EasyQuick RT MasterMix试剂盒(CW2019M,北京康为世纪生物科技有限公司)进行cDNA合成。采用Primer Premier 5软件设计荧光定量引物,引物序列为F:GGAACG CCCAACGTGCTGGACAA,R:CACCTGGATGC GTCCCATCTT;以玉米GAPDH为内参基因。利用Bio-Rad iQ5荧光定量PCR仪(Bio-RAD)及2×Fast Super EvaGreen® qPCR Mastermix试剂盒(BN12008,US Everbright Inc.)进行荧光定量分析。反应体系为:cDNA模板1 µL、前后引物各1 µL、ddH₂O 7 µL、2×AugeGreen™ Master Mix10 µL。基因相对表达量采用2⁻ΔΔCT法计算。
对培养约25 d的拟南芥进行控水处理,选取长势一致的WT和转基因株系植株,分为正常浇水对照组(CK)和干旱胁迫处理组(DR),DR处理组停止浇水10 d,观察表型变化并记录存活植株数量。同时取样进行生理指标测定,设3次生物学重复。使用BC0090、BC5160和BC0025试剂盒(北京索莱宝科技有限公司)分别测定POD活性、超氧化物歧化酶(SOD)活性和丙二醛(MDA)含量。
2 结果与分析
2.1 ZmPOD基因的克隆
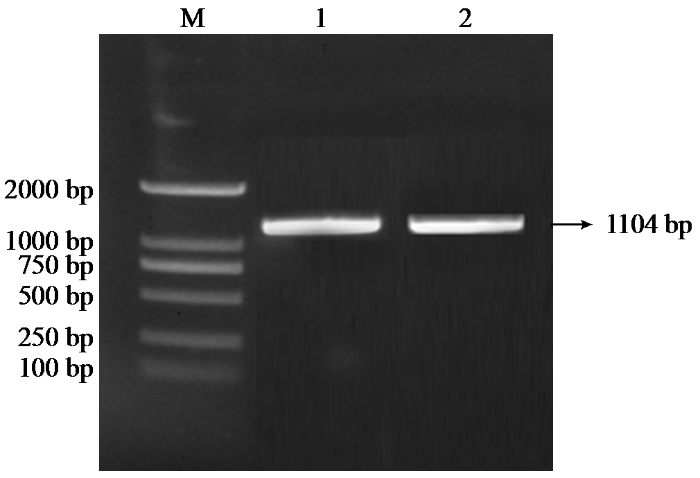
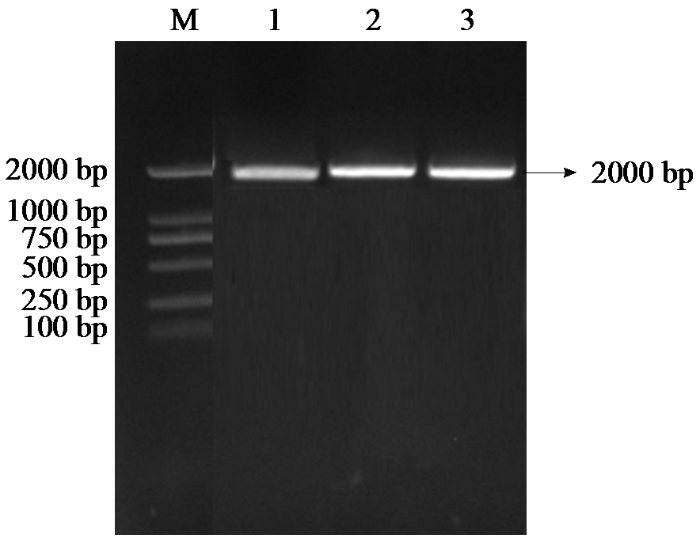
以玉米自交系农系3435的cDNA为模板,通过PCR扩增了ZmPOD基因的cDNA全长。经1%琼脂糖凝胶电泳检测(图1),扩增产物长度约为1104 bp,与ZmPOD基因的预期编码序列长度一致。将该片段回收纯化后进行测序,结果证实所克隆片段为ZmPOD基因的完整编码序列。
图1
2.2 ZmPOD蛋白质结构分析
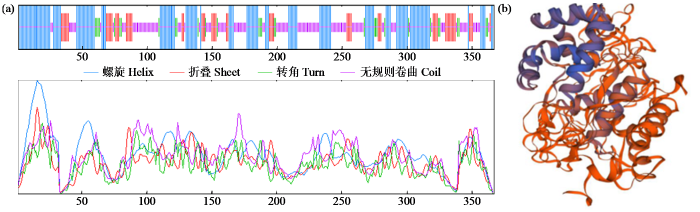
对ZmPOD蛋白序列进行分析,结果表明ZmPOD编码蛋白的分子式为C1672H2679N471O525S18,理论分子量为38.36 kDa,编码367个氨基酸(amino acid,AA),理论等电点为6.81,带负电荷的残基总数(Asp+Glu)为30,带正电荷的残基总数(Arg+ Lys)为30。
图2
图2
ZmPOD蛋白的二级结构(a)和三级结构(b)预测
Fig.2
The secondary structure (a) and tertiary structure (b) prediction of ZmPOD protein
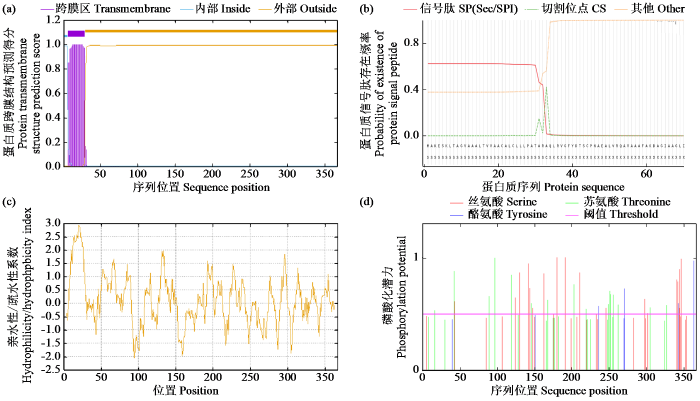
2.3 ZmPOD蛋白质序列分析
图3
2.4 ZmPOD蛋白序列比对
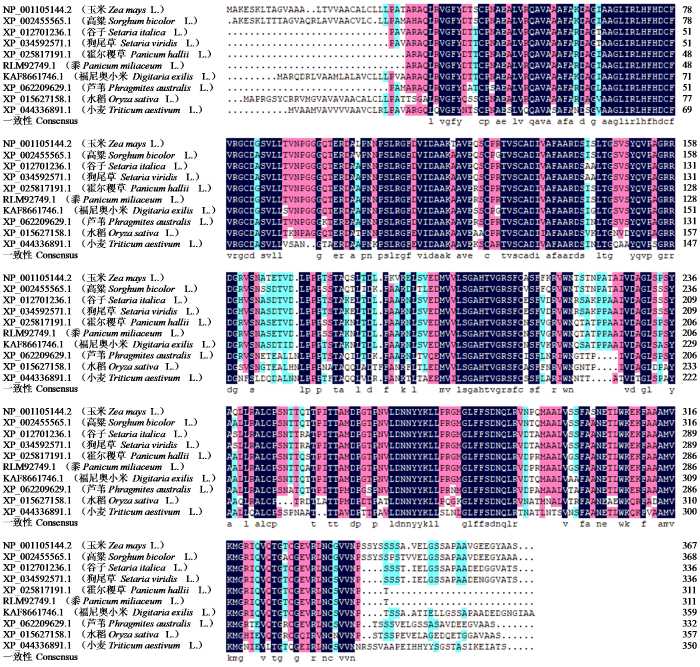
对ZmPOD在高粱、水稻、小麦、霍尔稷草、狗尾草、黍、谷子、福尼奥小米及芦苇中的蛋白序列进行多重比对分析,结果显示ZmPOD在这些物种中存在高度保守区域(图4)。同源性分析进一步表明,ZmPOD与上述物种具有较高的同源性,说明ZmPOD基因在不同物种间具有较高的保守特性。
图4
图4
不同植物ZmPOD蛋白序列比对
Fig.4
Sequence alignment of ZmPOD proteins in different plants
2.5 ZmPOD基因的启动子克隆与序列分析
图5
图6
图6
ZmPOD基因启动子顺式作用元件分析
Fig.6
Analysis of cis-acting elements of ZmPOD gene promoter
2.6 ZmPOD过表达拟南芥萌发期抗旱性鉴定
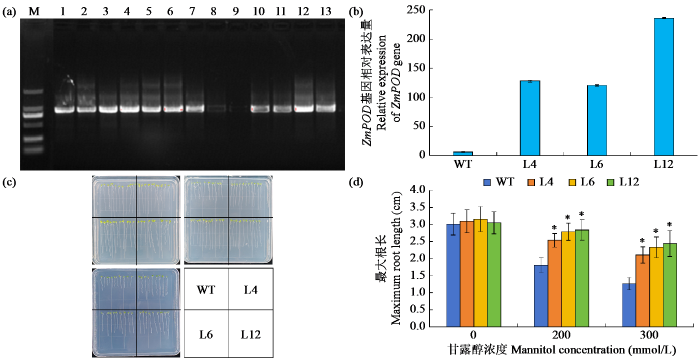
为验证ZmPOD基因的功能,选取ZmPOD基因表达量较高的T3代过表达株系L4、L6和L12(图7a,b)进行萌发期抗旱性鉴定。结果显示,在不含甘露醇的培养基中,WT与各转基因株系的根长无显著差异(图7c,d)。在加入甘露醇的培养基中,所有株系的生长均受到抑制,在含200 mmol/L甘露醇的1/2 MS培养基中,WT、L4、L6和L12株系的平均根长分别为1.81、2.54、2.79和2.84 cm;在300 mmol/L甘露醇条件下,平均根长分别为1.27、2.11、2.32和2.44 cm。综上所述,在干旱胁迫处理下,ZmPOD过表达株系的平均根长显著长于WT,证实ZmPOD基因在拟南芥中过表达能够有效增强幼苗期对干旱胁迫的耐受性。
图7
图7
ZmPOD过表达拟南芥萌发期抗旱性鉴定
(a)转基因拟南芥阳性植株筛选;(c)WT和过表达株系在不同浓度甘露醇中的生长情况。“*”表示与WT差异显著(P < 0.05)。
Fig.7
Drought-resistance identification of ZmPOD overexpression Arabidopsis at germination stage
(a) screening of transgenic Arabidopsis positive plants; (c) growth of WT and overexpressing seedlings at different concentrations of mannitol.“*”indicates significant difference from WT (P < 0.05).
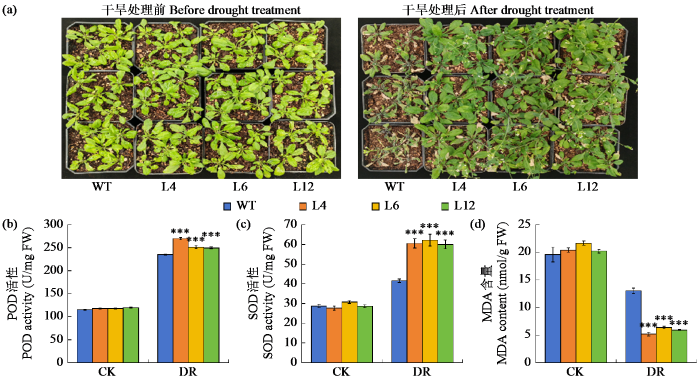
2.7 ZmPOD过表达拟南芥苗期抗旱性鉴定
图8
图8
ZmPOD过表达拟南芥苗期抗旱性鉴定
“***”表示与WT差异极显著(P < 0.001)。
Fig.8
Drought-resistance identification of ZmPOD overexpression Arabidopsis at seedling stage
“***”indicates extremely significant difference from WT (P < 0.001).
3 讨论
玉米作为重要作物,在生长过程中经常受到干旱等非生物胁迫影响,严重威胁其生产力与粮食安全。全球范围内,干旱导致的作物减产幅度超过其他因素造成减产的总和[3]。干旱胁迫对玉米的影响贯穿其生长的各个时期,涵盖从形态到分子水平的多个层面。玉米对干旱胁迫的响应属于数量性状,涉及复杂的基因调控网络。因此,挖掘玉米响应干旱胁迫的关键基因并培育耐旱玉米新品种,对于提升玉米耐旱能力具有重大意义。干旱胁迫会对植物体内多种生理生化过程产生不同程度的影响,包括植物生长发育、物质代谢、信号传递以及酶系统等方面。POD在植物应对干旱胁迫的反应中发挥重要作用,但其作用机制尚不明确。本研究成功克隆了玉米响应干旱胁迫的候选基因ZmPOD,利用生物信息学方法对其编码蛋白的结构进行分析与预测。结果表明,ZmPOD为不稳定的疏水性蛋白;氨基酸序列同源比对分析显示,ZmPOD与其他物种具有较高的相似性,推测ZmPOD基因编码的氨基酸结构域保守性较高。
近年来,生物技术的发展十分迅速,启动子研究已成为热点。深入探究启动子的结构与功能,有助于解析基因在转录和表达层面的调控机制。顺式调控元件的类型和数量,对决定基因的应激反应特性或植物组织特异性表达模式起着关键作用[21]。ZmPOD的上游启动子序列包含脱落酸响应元件(ABRE)、脱水响应元件(DRE core)、干旱诱导的MYB结合位点(MBS)、MYC元件、茉莉酸反应元件(CGTCA-motif)及与生物和非生物胁迫相关的响应元件(as-1)。据此推测,ZmPOD很可能受转录因子诱导而表达,并在水分胁迫过程中与特定转录因子互作以应对逆境。
为明确基因功能,常将目标基因转入模式作物,通过观察模式作物在胁迫条件下的表型及生理生化差异,对基因功能进行分析与验证[22]。拟南芥因具有植株矮小、繁殖周期短及生命力强等特性,成为模式作物的首选。本研究将ZmPOD基因转入拟南芥后发现,在甘露醇胁迫下,转基因株系的根长显著长于野生型;幼苗期经干旱处理后,转基因植株的生长状态优于野生型拟南芥,这表明ZmPOD基因增强了拟南芥的抗旱性。SOD和POD可通过清除植物体内多余的ROS,维持机体氧化平衡,保护植物免受损伤,因而常被用作衡量胁迫程度的关键指标[23]。干旱胁迫处理后,ZmPOD过表达株系的SOD和POD活性显著高于野生型,说明转基因株系能产生更多抗氧化物酶以抵御水分胁迫的危害。植物体内的自由基会促进脂质氧化,最终生成MDA,导致植物细胞产生毒性,加剧细胞膜损伤,因此MDA水平常作为植物抗性研究的关键指标[24]。ZmPOD过表达株系在干旱胁迫后,体内MDA含量增幅小于野生型材料,表明转基因株系耐旱性更强。综上所述,ZmPOD过表达拟南芥通过调节SOD活性、POD活性和MDA含量,减轻了干旱胁迫对拟南芥叶片细胞膜的损伤,进而提高了拟南芥的抗旱能力。综上所述,本研究通过生物信息学分析及ZmPOD过表达拟南芥植株试验,验证了该基因在干旱条件下的功能,为玉米抗旱基因的挖掘与利用提供了重要依据。
4 结论
基于前期多组学分析结果,成功从玉米中克隆出耐旱候选基因ZmPOD,该基因全长1104 bp,编码367个氨基酸;二级结构预测显示,α-螺旋与无规则卷曲占比最高。蛋白质序列分析表明,ZmPOD为含信号肽的跨膜疏水性蛋白。多序列比对结果显示,ZmPOD具有高度保守性,与高粱中的POD蛋白同源性最高;其启动子序列包含参与脱落酸、环境胁迫及干旱响应的顺式作用元件。在萌发期和幼苗期,ZmPOD基因过表达拟南芥株系的耐旱性均优于野生型植株,表明ZmPOD基因可提升拟南芥的耐旱能力。
参考文献
Drought Stress in Maize (Zea mays L.)
A systems approach to a spatio-temporal understanding of the drought stress response in maize
DOI:10.1038/s41598-017-04863-7
[本文引用: 1]

A correction to this article has been published and is linked from the HTML version of this paper. The error has been fixed in the paper.
Extreme climatic events: impacts of drought and high temperature on physiological processes in agronomically important plants
The class III peroxidase multigenic family in rice and its evolution in land plants
DOI:10.1016/j.phytochem.2004.06.023
PMID:15279994
[本文引用: 1]

Plant peroxidases (class III peroxidases, E.C. 1.11.1.7) are secreted glycoproteins known to be involved in the mechanism of cell elongation, in cell wall construction and differentiation, and in the defense against pathogens. They usually form large multigenic families in angiosperms. The recent completion of rice (Oryza sativa japonica c.v. Nipponbare) genome sequencing allowed drawing up the full inventory of the genes encoding class III peroxidases in this plant. We found 138 peroxidase genes distributed among the 12 rice chromosomes. In contrast to several other gene families studied so far, peroxidase genes are twice as numerous in rice as in Arabidopsis. This large number of genes results from various duplication events that were tentatively traced back using a phylogenetic tree based on the alignment of conserved amino acid sequences. We also searched for peroxidase encoding genes in the major phyla of plant kingdom. In addition to gymnosperms and angiosperms, sequences were found in liverworts, mosses and ferns, but not in unicellular green algae. Two rice and one Arabidopsis peroxidase genes appeared to be rather close to the only known sequence from the liverwort Marchantia polymorpha. The possible relationship of these peroxidases with the putative ancestor of peroxidase genes is discussed, as well as the connection between the development of the class III peroxidase multigenic family and the emergence of the first land plants.
A large family of class III plant peroxidases
Plant peroxidases: structure function relationships
Crystal structure of lignin peroxidase
Probing the structure and bifunctionality of catalase-peroxidase (KatG)
PMID:16516299

Catalase-peroxidases (KatGs) exhibit peroxidase and substantial catalase activities similar to monofunctional catalases. Crystal structures of four different KatGs reveal the presence of a peroxidase-conserved proximal and distal heme pocket together with features unique to KatG. To gain insight into their structure-function properties, many variants were produced and very similar results were obtained irrespective of the origin of the KatG mutated. This review focuses mainly on the electronic absorption and resonance Raman results together with the combined analysis of pre-steady and steady-state kinetics of various mutants involving both the peroxidase-conserved and the KatG-specific residues of recombinant KatG from the cyanobacterium Synechocystis. Marked differences in the structural role of conserved amino acids and hydrogen-bond networks in KatG with respect to the other plant peroxidases were found. Typically, the catalatic but not the peroxidatic activity was very sensitive to mutations that disrupted the KatG-typical extensive hydrogen-bonding network. Moreover, the integrity of this network is crucial for the formation of distinct protein radicals formed upon incubation of KatG with peroxides in the absence of one-electron donors. The correlation between the structural architecture and the bifunctional activity is discussed and compared with data obtained for KatGs from other organisms.
Prokaryotic origins of the non-animal peroxidase superfamily and organelle-mediated transmission to eukaryotes
Members of the superfamily of plant, fungal, and bacterial peroxidases are known to be present in a wide variety of living organisms. Extensive searching within sequencing projects identified organisms containing sequences of this superfamily. Class I peroxidases, cytochrome c peroxidase (CcP), ascorbate peroxidase (APx), and catalase peroxidase (CP), are known to be present in bacteria, fungi, and plants, but have now been found in various protists. CcP sequences were detected in most mitochondria-possessing organisms except for green plants, which possess only ascorbate peroxidases. APx sequences had previously been observed only in green plants but were also found in chloroplastic protists, which acquired chloroplasts by secondary endosymbiosis. CP sequences that are known to be present in prokaryotes and in Ascomycetes were also detected in some Basidiomycetes and occasionally in some protists. Class II peroxidases are involved in lignin biodegradation and are found only in the Homobasidiomycetes. In fact class II peroxidases were identified in only three orders, although degenerate forms were found in different Pezizomycota orders. Class III peroxidases are specific for higher plants, and their evolution is thought to be related to the emergence of the land plants. We have found, however, that class III peroxidases are present in some green algae, which predate land colonization. The presence of peroxidases in all major phyla (except vertebrates) makes them powerful marker genes for understanding the early evolutionary events that led to the appearance of the ancestors of each eukaryotic group.
Cytochrome c in the apoptotic and antioxidant cascades
DOI:10.1016/s0014-5793(98)00061-1
PMID:9515723
[本文引用: 1]

Recent progress in studies on apoptosis has revealed that cytochrome c is a pro-apoptotic factor. It is released from its places on the outer surface of the inner mitochondrial membrane at early steps of apoptosis and, combining with some cytosolic proteins, activates conversion of the latent apoptosis-promoting protease pro-caspase-9 to its active form. Cytochrome c release can be initiated by the pro-apoptotic protein Bax. This process is blocked by the anti-apoptotic proteins Bcl-2 and Bcl-xL. The role of cytochrome c in apoptosis may be understood within the framework of the concept assuming that the evolutionary primary function of apoptosis was to purify tissues from ROS-overproducing cells. In this context, the pro-apoptosis activity of cytochrome c might represent one of the anti-oxidant functions inherent in this cytochrome. Among other cytochrome c-linked antioxidant mechanisms, the following systems can be indicated. (1) Cytochrome c released from the inner mitochondrial membrane to the intermembrane space can operate as an enzyme oxidizing O2.- back to O2. The reduced cytochrome c is oxidized by cytochrome oxidase (or in yeasts and bacteria, by cytochrome c peroxidase). (2) The intermembrane cytochrome c can activate the electron transport chain in the outer mitochondrial membrane. This bypasses the initial and middle parts of the main respiratory chain, which produce, as a rule, the major portion of ROS in the cell. (3) The main respiratory chain losing its cytochrome c is inhibited in such a fashion that antimycin-like agents fail to stimulate ROS production.
Yeast cytochrome c peroxidase: mechanistic studies via protein engineering
A new versatile peroxidase from Pleurotus
DOI:10.1042/bst0290116 URL [本文引用: 1]
Specific functions of individual class III peroxidase genes
DOI:10.1093/jxb/ern318
PMID:19088338
[本文引用: 1]

In higher plants, class III peroxidases exist as large multigene families (e.g. 73 genes in Arabidopsis thaliana). The diversity of processes catalysed by peroxidases as well as the large number of their genes suggests the possibility of a functional specialization of each isoform. In addition, the fact that peroxidase promoter sequences are very divergent and that protein sequences contain both highly conserved domains and variable regions supports this hypothesis. However, two difficulties are associated with the study of the function of specific peroxidase genes: (i) the modification of the expression of a single peroxidase gene often results in no visible mutant phenotype, because it is compensated by redundant genes; and (ii) peroxidases show low substrate specificity in vitro resulting in an unreliable indication of peroxidase specific activity unless complementary data are available. The generalization of molecular biology approaches such as whole transcriptome analysis and recombinant DNA combined with biochemical approaches provide unprecedented tools for overcoming these difficulties. This review highlights progress made with these new techniques for identifying the specific function of individual class III peroxidase genes taking as an example the model plant A. thaliana, as well as discussing some other plants.
A mechanistic study on the toxic effect of copper oxide nanoparticles in soybean (Glycine max L.) root development and lignification of root cells
DOI:10.1007/s12011-014-0106-5 URL [本文引用: 1]
Peroxidase- dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance
DOI:10.1111/tpj.2006.47.issue-6 URL [本文引用: 1]
Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC tran scription factor family in rice
DOI:10.1007/s00438-008-0386-6 URL [本文引用: 1]