作物杂志,2021, 第4期: 10–17 doi: 10.16035/j.issn.1001-7283.2021.04.002
燕麦属植物核糖体DNA染色体定位及45S rDNA的系统进化分析
王炳策1( ), 刘晓娟1(
), 刘晓娟1( ), 程斌2, 任明见1, 徐如宏1, 张素勤1, 张立异2, 何方1(
), 程斌2, 任明见1, 徐如宏1, 张素勤1, 张立异2, 何方1( )
)
- 1贵州大学农学院/国家小麦改良中心贵州分中心,550025,贵州贵阳
2贵州省农业科学院旱粮研究所,550006,贵州贵阳
Chromosomal Localization of Ribosomal DNA and Phylogenetic Analysis of 45S rDNA in Avena
Wang Bingce1( ), Liu Xiaojuan1(
), Liu Xiaojuan1( ), Cheng Bin2, Ren Mingjian1, Xu Ruhong1, Zhang Suqin1, Zhang Liyi2, He Fang1(
), Cheng Bin2, Ren Mingjian1, Xu Ruhong1, Zhang Suqin1, Zhang Liyi2, He Fang1( )
)
- 1College of Agriculture, Guizhou University/Guizhou Subcenter of National Wheat Improvement Center, Guiyang 550025, Guizhou, China
2Institute of Upland Crops, Guizhou Academy of Agriculture Sciences, Guiyang 550006, Guizhou, China
摘要:
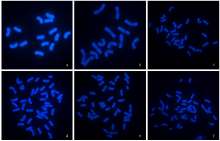
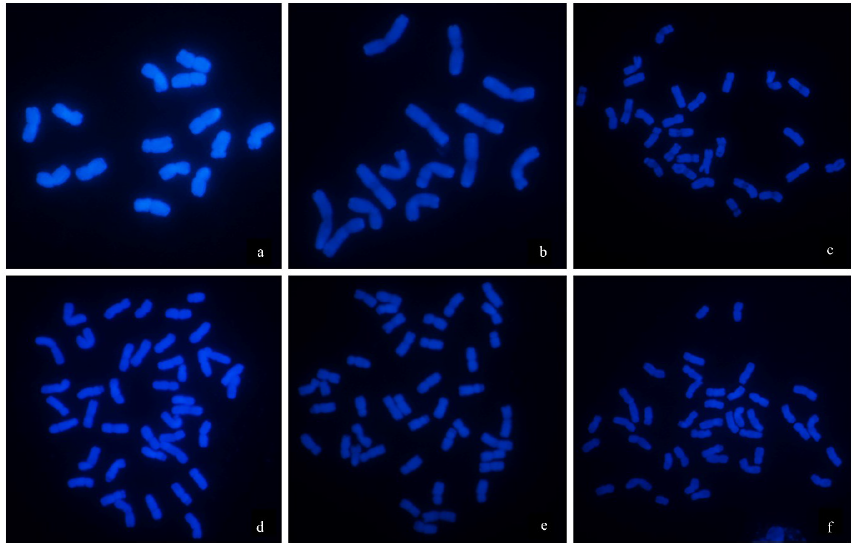
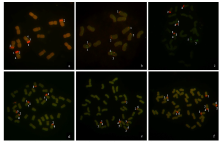
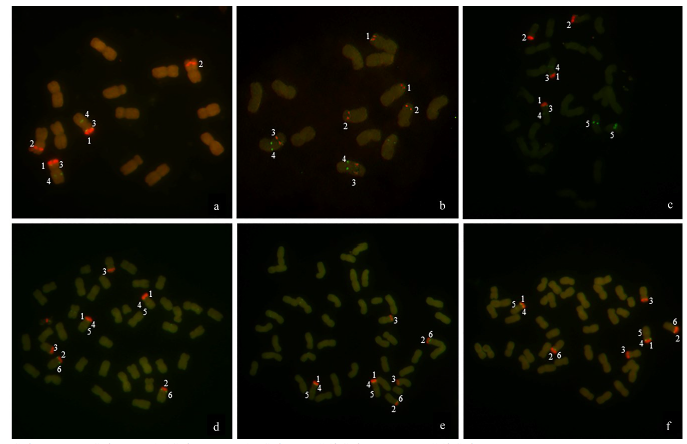
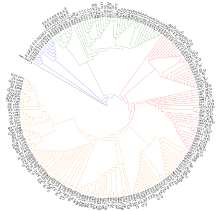
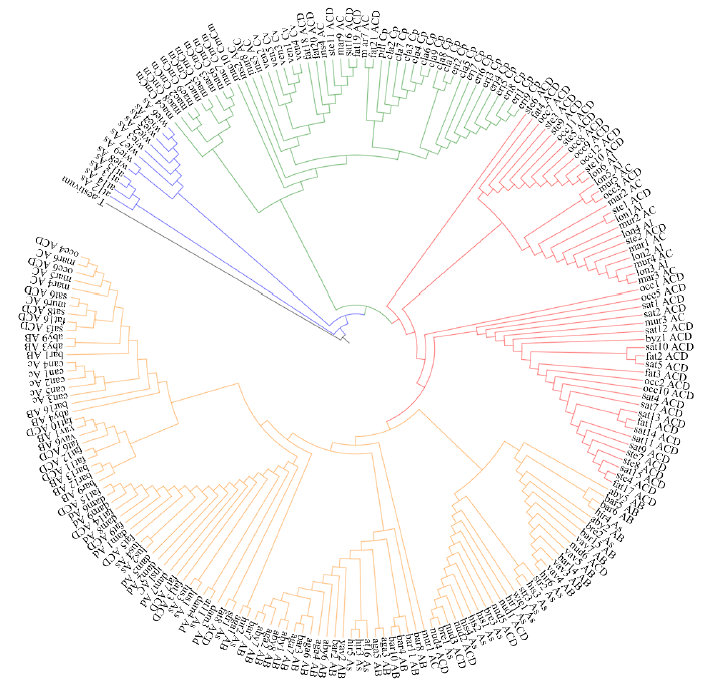
由于缺乏明确的二倍体供体信息,燕麦属植物的起源和系统进化关系一直存在争议。利用荧光原位杂交(fluorescence in situ hybridization,FISH)方法,检测45S rDNA和5S rDNA在燕麦属不同倍性植物染色体上的位点信息;并依据已公开的45S rDNA ITS区全长DNA序列构建分子进化树。探讨燕麦属植物在不同基因组中45S rDNA的位点变化、进化规律以及分化机制,为探究燕麦属物种的起源与演化提供参考。
| [1] | 李威, 周青平. 六种裸燕麦品种种子萌发期抗旱性的研究. 草业与畜牧, 2008,5(9):5-8. |
| [2] | 彭远英, 颜红海, 郭来春, 等. 燕麦属不同倍性种质资源抗旱性状评价及筛选. 生态学报, 2011,31(9):134-147. |
| [3] | 李英丽, 方正, 毛明艳. 不同燕麦品种耐碱性筛选和鉴定. 河北农业大学学报, 2014,37(6):13-17. |
| [4] | 董玉琛, 刘旭总. 中国作物及其野生近缘植物:粮食作物卷. 北京: 中国农业出版社, 2006. |
| [5] | Drossou A, Katsiotis A, Leggett J M, et al. Genome and species relationships in genus Avena based on RAPD and AFLP molecular markers. Theoretical and Applied Genetics, 2004,109(1):48-54. |
| [6] | Badaeva E D, Loskutov I G, Shelukhina O Y, et al. Cytogenetic analysis of diploid Avena L. species containing the as genome. Russian Journal of Genetics, 2005,41(12):1428-1433. |
| [7] | Rodionov A V, Tyupa N B, Kim E S, et al. Genomic configuration of the autotetraploid oat species avena macrostachya inferred from comparative analysis of ITS1 and ITS2 sequences:on the oat karyotype evolution during the early events of the Avena species divergence. Russian Journal of Genetics, 2005,41(5):518-528. |
| [8] | Loskutov I G. On evolutionary pathways of Avena species. Genetic Resources and Crop Evolution, 2008,55(2):211-220. |
| [9] | Nikoloudakis N, Skaracis G, Katsiotis A. Evolutionary insights inferred by molecular analysis of the ITS1-5.8S-ITS2 and IGS Avena sp. sequences. Molecular Phylogenetics and Evolution, 2008,46(1):102-115. |
| [10] | 刘青, 刘欢, 林磊. 燕麦属系统学研究进展. 热带亚热带植物学报, 2014(5):516-524. |
| [11] | 龚志云, 吴信淦, 程祝宽, 等. 水稻45S rDNA和5S rDNA的染色体定位研究. 遗传学报, 2002,29(3):241-244. |
| [12] | Pontes O, Cotrim H, Pais S, et al. Physical mapping,expression patterns and interphase organisation of rDNA loci in Portuguese endemic Silene cintrana and Silene rothmaleri. Chromosome Research, 2000,8(4):313-317. |
| [13] | Kato A, Lamb J C, Birchler J A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proceedings of the National Academy of Sciences, 2004,101(37):13554-13559. |
| [14] | Mahelka V, Kopecký D, Baum B R. Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Molecular Biology and Evolution, 2013,30(9):2065-2086. |
| [15] | Clegg M T, Zurawski G. Chloroplast DNA and the study of plant phylogeny:present status and future prospects//Soltis P S,Soltis D E,Doyle J J. Molecular Systematics of Plants. Springer:Boston,MA, 1992. |
| [16] | Peng Y Y, Wei Y M, Baum B R, et al. Molecular diversity of the 5S rRNA gene and genomic relationships in the genus Avena (Poaceae:Aveneae). Genome, 2008,51(2):137-154. |
| [17] | Yan H H, Baum B R, Zhou P P, et al. Phylogenetic analysis of the genus Avena based on chloroplast intergenic spacer psb A-trn H and single-copy nuclear gene Acc1. Genome, 2014,57(5):267-277. |
| [18] | Fu Y B. Oat evolution revealed in the maternal lineages of 25 Avena species. Scientific Reports, 2018,8(1):1-12. |
| [19] | Thompson J D, Gibson T J, Plewniak F, et al. The CLUSTAL_X Windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 1997,25(24):4876-4882. |
| [20] | Kumar S, Stecher G, Li M, et al. MEGA X:Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 2018,35(6):1547-1549. |
| [21] | Linc G, Gaál E, Molnár I, et al. Molecular cytogenetic (FISH) and genome analysis of diploid wheatgrasses and their phylogenetic relationship. PLoS ONE, 2017,12(3):e0173623. |
| [22] | Mahelka V, Kopecký D, Baum B R. Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Molecular Biology and Evolution, 2013,30(9):2065-2086. |
| [23] | 赵筱芳. 冰草居群45S rDNA基因位点FISH分析. 成都:四川农业大学, 2016. |
| [24] | Choi H W, Koo D H, Bang K H, et al. FISH and GISH analysis of the genomic relationships among Panax species. Genes and Genomics, 2009,31(1):99-105. |
| [25] | 符文炎, 刘义飞, 黄宏文. 荧光原位杂交技术在植物多倍体起源与进化研究中的应用. 热带亚热带植物学报, 2014,22(3):314-322. |
| [26] | Manns U, Anderberg A A. Molecular phylogeny of Anagallis (Myrsinaceae) based on ITS, trn L-F,and ndh F sequence data. International Journal of Plant Sciences, 2005,166(6):1019-1028. |
| [27] | Gillespie L J, Soreng R J, Bull R D, et al. Phylogenetic relationships in subtribe Poinae (Poaceae,Poeae) based on nuclear ITS and plastid trn T-trn L-trn F sequences. Botany, 2008,86(8):938-967. |
| [28] | Attar F, Riahi M, Daemi F, et al. Preliminary molecular phylogeny of Eurasian Scrophularia (Scrophulariaceae) based on DNA sequence data from trnS-trnG and ITS regions. Plant Biosystems, 2011,145(4):857-865. |
| [1] | 韩兴,何运转,康占海,王高杨,王冰,李亚宁,刘大群. 掌叶半夏茎基腐病病原鉴定及其防治药剂筛选[J]. 作物杂志, 2019, (6): 177–181 |
| [2] | 李凤兰, 蒋先锋, 史丽娟, 等. 两株马铃薯干腐病病原菌的分离和鉴定[J]. 作物杂志, 2013, (4): 125–128 |
|
||