作物杂志,2020, 第5期: 23–32 doi: 10.16035/j.issn.1001-7283.2020.05.004
亚麻荠油脂相关转录因子CsLEC2基因家族的鉴定及表达分析
王志龙1( ), 薛应红1, 郝月茹1, 刘宝玲1, 苑丽霞2, 薛金爱1(
), 薛应红1, 郝月茹1, 刘宝玲1, 苑丽霞2, 薛金爱1( ), 李润植1
), 李润植1
- 1山西农业大学分子农业与生物能源研究所,030801,山西太谷
2晋中学院生物科学与技术学院,030600,山西榆次
Identification and Expression Analysis of Oil-Related Transcription actor CsLEC2 Gene Family in Camelina sativa (L.) Crantz
Wang Zhilong1( ), Xue Yinghong1, Hao Yueru1, Liu Baoling1, Yuan Lixia2, Xue Jin'ai1(
), Xue Yinghong1, Hao Yueru1, Liu Baoling1, Yuan Lixia2, Xue Jin'ai1( ), Li Runzhi1
), Li Runzhi1
- 1Institute of Molecular Agriculture and Bioenergy, Shanxi Agricultural University, Taigu 030801, Shanxi, China
2College of Biological Science and Technology, Jinzhong University, Yuci 030600, Shanxi, China
摘要:
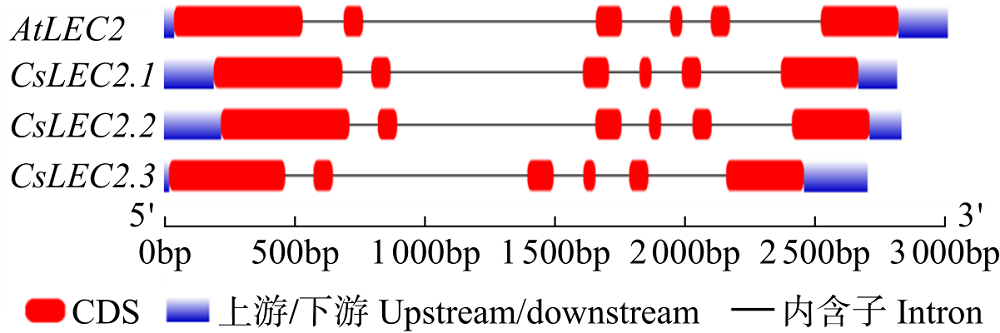
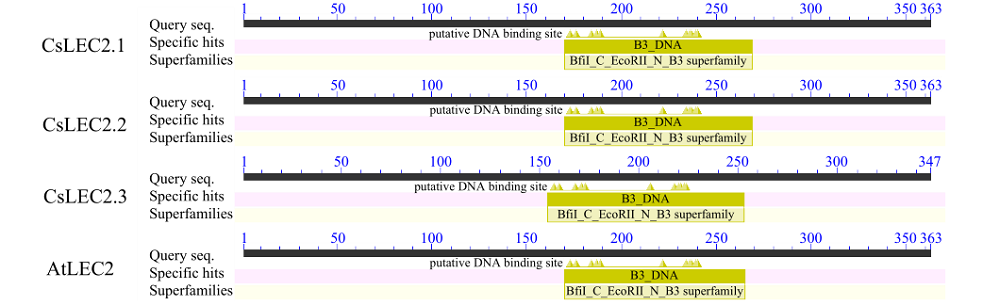
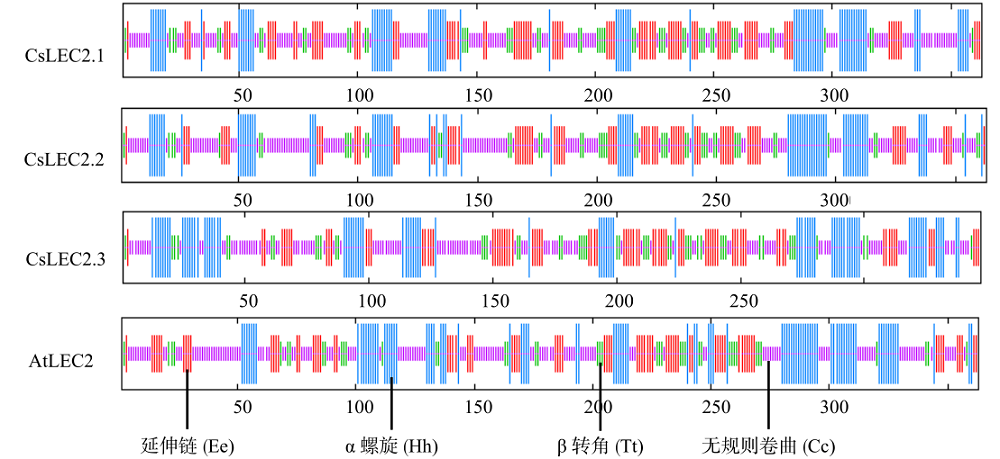
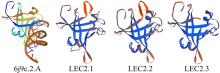
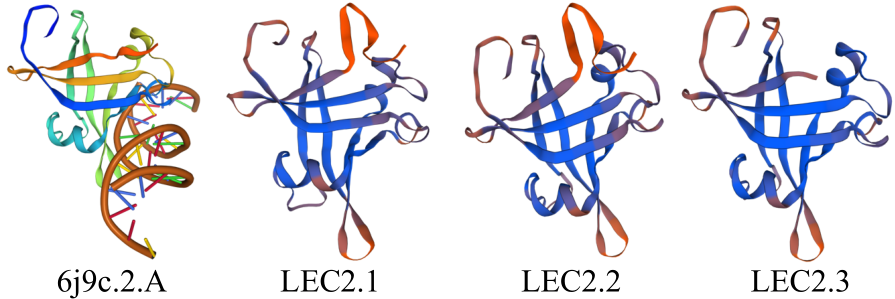
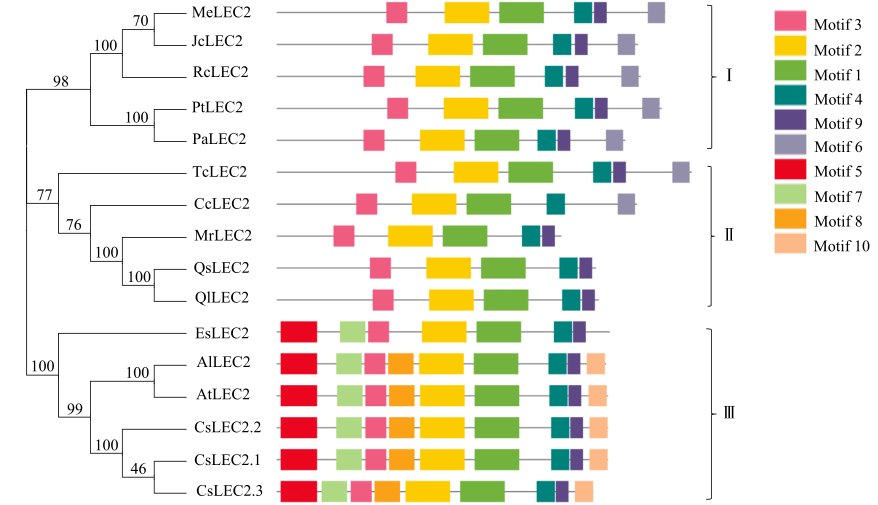
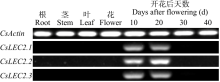
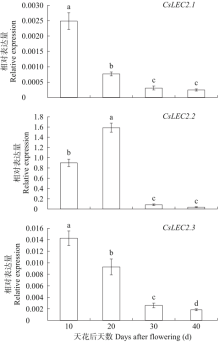
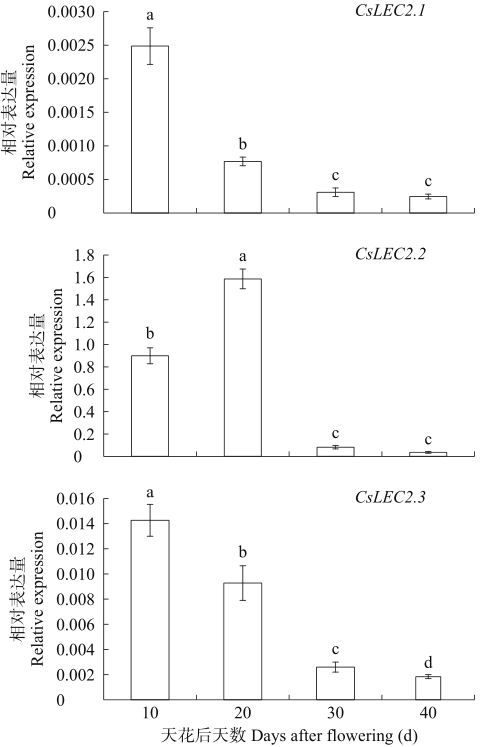
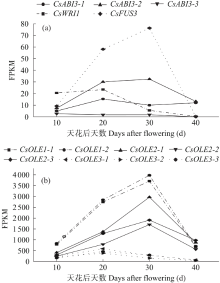
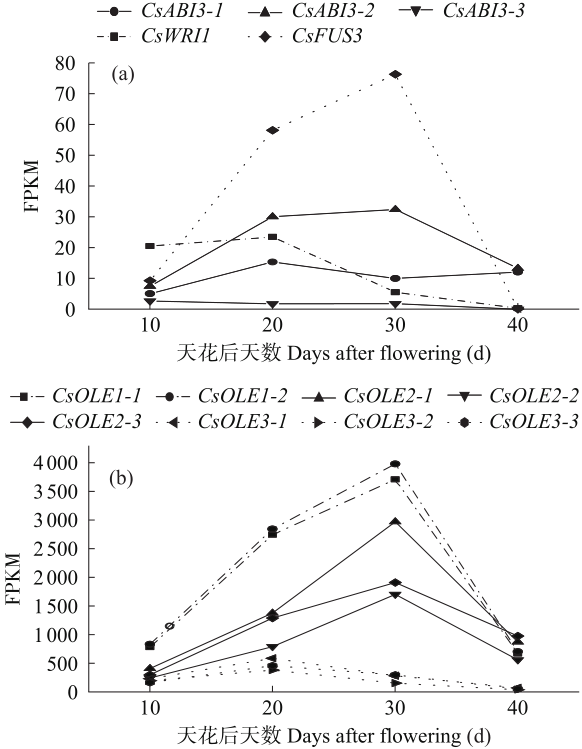
对亚麻荠CsLEC2家族进行全基因组鉴定、半定量RT-PCR、qRT-PCR,检测CsLEC2基因的时空表达特性并通过转录组数据预测CsLEC2下游靶基因,以解析CsLEC2调控亚麻荠种子油脂合成和积累等生物学功能。结果表明,在亚麻荠基因组中共鉴定到3个亚麻荠CsLEC2基因(CsLEC2.1、CsLEC2.2和CsLEC2.3)。蛋白理化性质和高级结构分析显示,亚麻荠CsLEC2蛋白具有与拟南芥AtLEC2相似的理化性质,且其二级结构的主体也相似。系统进化分析表明,亚麻荠CsLEC2蛋白与模式植物拟南芥AtLEC2蛋白及拟南芥琴亚种AlLEC2蛋白亲缘关系最近。qRT-PCR结果显示亚麻荠CsLEC2在未成熟的种子中高表达。转录组数据分析显示下游基因CsWRI1和CsOLE3高表达,推测CsLEC2可能直接上调CsWRI1和CsOLE3的转录表达,参与油脂代谢调控。
| [1] | 苑丽霞, 毛雪, 高昌勇 等. 新型工业油料作物亚麻荠:从基因组到代谢工程. 植物生理学报, 2015,51(8):1204-1216. |
| [2] |
Kang J L, Lu C F, et al. Identification of three genes encoding microsomal oleate desaturases (FAD2) from the oil seed crop Camelina sativa. Plant Physiology and Biochemistry, 2010,49(2011):223-229.
doi: 10.1016/j.plaphy.2010.12.004 |
| [3] |
Lu C F, Kang J L. Generation of transgenic plants of a potential oil seed crop Camelina sativa by Agrobacterium-mediated transformation. Plant Cell Reports, 2008,27:273-278.
doi: 10.1007/s00299-007-0454-0 |
| [4] | 李昌珠, 李正茂. 植物脂肪酸的生物合成及其生理功能的研究进展. 湖南林业科技, 2009,36(6):45-49. |
| [5] |
Nakano T, Suzuki K, Fujimura T, et al. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology, 2006,140:411-432.
doi: 10.1104/pp.105.073783 pmid: 16407444 |
| [6] | 郭秀芬, 张海龙, 王明晶 等. LAFL基因在种子发育和萌发中的功能. 植物生理学报, 2019,55(4):393-400. |
| [7] |
Meinke D W. A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science, 1992,258(5088):1647-1650.
doi: 10.1126/science.258.5088.1647 pmid: 17742538 |
| [8] | Stone S L, Kwong L W, Yee K M, et al. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proceedings of the National Academy of Sciences of the United States of America, 2001,98(20):11806-11811. |
| [9] |
Wójcikowska B, Jaskóła K, Gąsiorek P, et al. LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis,via YUCCA-mediated auxin biosynthesis. Planta, 2013,238(3), 425-440.
doi: 10.1007/s00425-013-1892-2 |
| [10] |
Swaminathan K, Peterson K, Jack T. The plant B3 superfamily. Trends in Plant Science, 2008,13(12), 647-655.
doi: 10.1016/j.tplants.2008.09.006 |
| [11] |
Baud S, Kelemen Z, Thévenin J, et al. Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by L-AFL proteins in Arabidopsis seed. Plant Physiology, 2016,171:1099-1121.
doi: 10.1104/pp.16.00034 pmid: 27208266 |
| [12] | 唐通. 植物中油脂调节基因ABSCISIC ACID INSENSITIVE3、FUSCA3和LEAFY COTYLEDON2的鉴定、系统进化和结构特征分析. 杨凌:西北农林科技大学, 2019. |
| [13] |
Mendoza M S, Dubreucq B, Miquel M, et al. LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Letters, 2005,579(21):4666-4670.
doi: 10.1016/j.febslet.2005.07.037 pmid: 16107256 |
| [14] |
Kim H U, Jung S J, Lee K R, et al. Ectopic overexpression of castor bean LEAFY COTYLEDON2 (LEC2) in Arabidopsis triggers the expression of genes that encode regulators of seed maturation and oil body proteins in vegetative tissues. FEBS Open Bio, 2014,4:25-32.
doi: 10.1016/j.fob.2013.11.003 pmid: 24363987 |
| [15] |
Baud S, Mendoza M S, Dubreucq B, et al. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. The Plant Journal, 2007,50(5), 825-838.
doi: 10.1111/j.1365-313X.2007.03092.x pmid: 17419836 |
| [16] | Braybrook S A, Stone S L, Park S, et al. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proceeding of the National Academy of Sciences of the United States of America, 2006,103(9):3468-3473. |
| [17] | Kroj T, Savino G, Valon C, et al. Regulation of storage protein gene expression in Arabidopsis. Development, 2003,130(24):60-73. |
| [18] |
Angeles-Núñez J G, Tiessen A. Mutation of the transcription factor LEAFY COTYLEDON 2 alters the chemical composition of Arabidopsis seeds,decreasing oil and protein content,while maintaining high levels of starch and sucrose in mature seeds. Journal of Plant Physiology, 2011,168(16), 1891-1900.
doi: 10.1016/j.jplph.2011.05.003 |
| [19] |
Kagale S, Nixon J, Khedikar Y, et al. The developmental transcriptome atlas of the biofuel crop Camelina Sativa. The Plant Journal, 2016,88(5), 879-894.
doi: 10.1111/tpj.13302 pmid: 27513981 |
| [20] |
Tao Z, Hu H M, Luo X, et al. Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nature Plants, 2019,5(4):424-435.
doi: 10.1038/s41477-019-0402-3 pmid: 30962525 |
| [21] |
Troncoso-Ponce M A, Barthole G, Tremblais G, et al. Transcriptional activation of two delta-9 palmitoyl-ACP desaturase genes by MYB115 and MYB118 is critical for biosynthesis of omega-7 monounsaturated fatty acids in the endosperm of Arabidopsis seeds. Plant Cell, 2016,28:2666-2682.
doi: 10.1105/tpc.16.00612 pmid: 27681170 |
| [22] |
Boulard C, Thévenin J, Tranquet O, et al. LEC1 (NF-YB9) directly interacts with LEC2 to control gene expression in seed. Biochimica et Biophysica Acta, 2018,1861(5):443-450.
doi: 10.1016/j.bbagrm.2018.03.005 pmid: 29580949 |
| [23] |
Bhardwaj J, Chauhan R, Swarnkar M K, et al. Comprehensive transcriptomic study on horse gram (Macrotyloma uniflorum):de novo assembly,functional characterization and comparative analysis in relation to drought stress. BMC Genomics, 2013,14:647-663.
doi: 10.1186/1471-2164-14-647 pmid: 24059455 |
| [24] | 杨英军, 周鹏. 番木瓜proteinase omega基因启动子的克隆及功能初步研究. 云南植物研究, 2005(5):99-105. |
| [25] | 果天宇, 尹悦佳, 贾伟刘, 等. 玉米ZmCOL3_(pro217)启动子的克隆及功能分析. 玉米科学, 2020,28(2):54-60. |
| [26] | 马彦龙. 大豆GmLEC2基因的克隆及再生功能的初步分析. 哈尔滨:东北农业大学, 2016. |
| [1] | 徐园园, 赵鹏, 洪权春, 朱晓琴, 裴冬丽. 小麦转录因子基因TaMYB70的分离和表达分析[J]. 作物杂志, 2020, (4): 84–90 |
| [2] | 宋健,晓宁,王海岗,陈凌,王君杰,刘思辰,乔治军. SiASRs家族基因的鉴定及表达分析[J]. 作物杂志, 2019, (6): 33–42 |
| [3] | 李钰,郑文寅,冯春,王荣富,李娟. 非生物逆境胁迫下普通小麦烟农19幼苗FeSOD基因表达分析[J]. 作物杂志, 2016, (4): 75–79 |
| [4] | 袁红梅,郭文栋,赵丽娟,于莹,吴建忠,程莉莉,赵东升,康庆华,黄文功,姚玉波,宋喜霞,姜卫东,刘岩,马廷芬,吴广文,关凤芝. 亚麻糖基转移酶基因LuUGT72E1的克隆与表达分析[J]. 作物杂志, 2016, (4): 62–67 |
| [5] | 向鹏, 龙承波, 罗红丽. 水稻OsMED7基因的克隆及表达分析[J]. 作物杂志, 2013, (3): 21–24 |
|
||