Crops ›› 2016, Vol. 32 ›› Issue (4): 62-67.doi: 10.16035/j.issn.1001-7283.2016.04.010
Previous Articles Next Articles
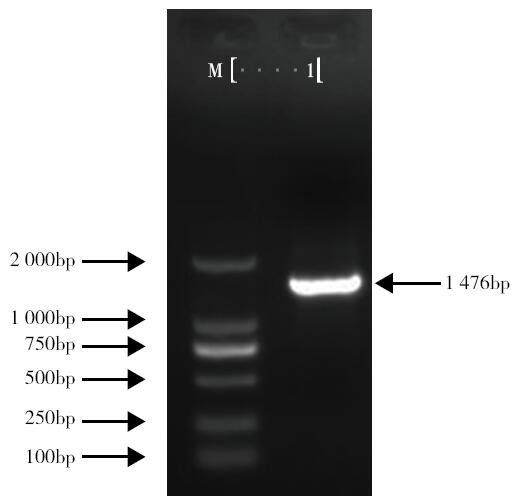
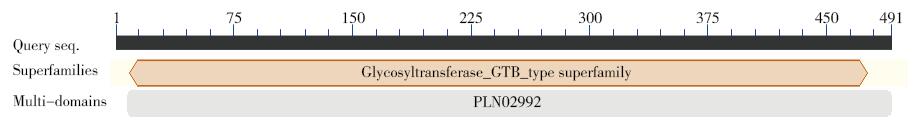
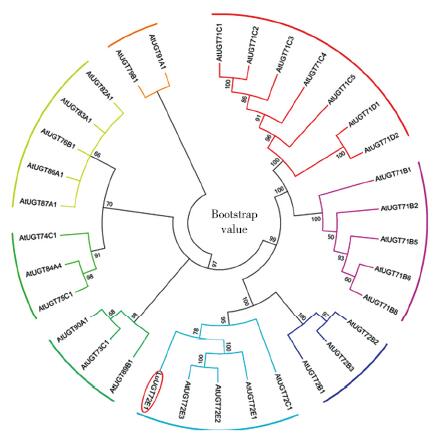
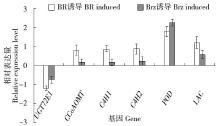
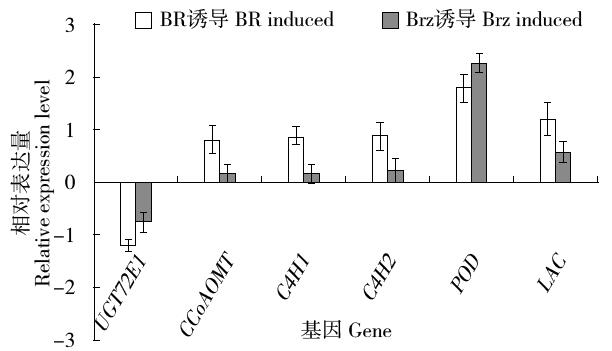
Cloning and Expression Analysis of the Glycosyltransferase Gene LuUGT72E1 in Flax
Yuan Hongmei1,2,Guo Wendong3,Zhao Lijuan4,Yu Ying2,Wu Jianzhong2,Cheng Lili2,Zhao Dongsheng2,Kang Qinghua2,Huang Wengong2,Yao Yubo2,Song Xixia2,Jiang Weidong2,Liu Yan2,Ma Tingfen2,Wu Guangwen2,Guan Fengzhi2
- 1 Heilongjiang Academy of Agricultural Sciences Postdoctoral Programme,Harbin 150086,Heilongjiang,China
2 Industrial Crops Institute,Heilongjiang Academy of Agricultural Sciences,Harbin 150086,Heilongjiang,China
3 Nature and Ecology Institute,Heilongjiang Academy of Sciences,Harbin 150040,Heilongjiang,China
4 Crop Breeding Institute,Heilongjiang Academy of Agricultural Sciences,Harbin 150086,Heilongjiang,China
| [1] | Franke R, Hemm M R, Denault J W , et al. Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Planta, 2002,30(1):47-59. |
| [2] |
Novaes E, Kirst M, Chiang V , et al. Lignin and biomass:a negative correlation for wood formation and lignin content in trees. Plant Physiology, 2010,154(2):555-561.
doi: 10.1104/pp.110.161281 |
| [3] | 魏建华, 宋艳茹 . 木质素生物合成途径及调控的研究进展. 植物学报, 2001,43(8) : 771-779. |
| [4] |
陈晓光, 史春余, 尹燕枰 , 等. 小麦茎秆木质素代谢及其与抗倒性的关系. 作物学报, 2011,37(9):1616-1622.
doi: 10.3724/SP.J.1006.2011.01616 |
| [5] | Jones L, Ennos A R, Turner S R . Cloning and characterization of irregular xylem4 (irx4):A severely lignin-deficient mutant of Arabidopsis. Planta, 2001,26(2):205-216. |
| [6] |
Ruben V, Véronique S, Bartel V , et al. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell, 2012,24(9):3506-3529.
doi: 10.1105/tpc.112.102574 |
| [7] |
Marie B, Bernard M, Marc V M , et al. Biosynthesis and genetic enginering of lignin. Critical Reviews in Plant Sciences, 1998,17(2):125-197.
doi: 10.1080/07352689891304203 |
| [8] |
Barry H . Lignin synthesis:the generation of hydrogen peroxide and superoxide by horseradish peroxidase and its stimulation by manganese(II) and phenols. Planta, 1978,140(1):81-88.
doi: 10.1007/BF00389384 |
| [9] | Du X, Gellerstedt G, Li J . Universal fractionation of lignin-carbohydrate complexes(LCCs) from lignocellulosic biomass:an example using spruce wood. Planta, 2013,74(2):328-338. |
| [10] |
Xu L, Nicholas D B, Jing K W , et al. The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. The Plant Cell, 2010,22(5):1620-1632.
doi: 10.1105/tpc.110.074161 |
| [11] | Du X Y, Marta P B, Carmen F , et al. Analysis of lignin-carbohydrate and lignin-lignin linkages after hydrolase treatment of xylan-lignin,glucomannan-lignin and glucan-lignin complexes from spruce wood. Planta, 2014,4(15):323-358. |
| [12] | Hao Z, Mohnen D . A review of xylan and lignin biosynthesis:Foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes.Critical Reviews Biochemistry Molecular Biology, 2014( doi:10.3109/10409238.2014.889651). |
| [13] | 王艳文 .杨树糖基转移酶与木质素合成关系的转基因研究. 济南:山东大学, 2012. |
| [14] | Lanot A, Hodge D, Jackson R G , et al. The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis. Planta, 2006,48(2):286-295. |
| [15] |
Lanot A, Hodge D, Lim E K , et al. Redirection of flux through the phenylpropanoid pathway by increased glucosylation of soluble intermediates. Planta, 2008,228(4):609-616.
doi: 10.1007/s00425-008-0763-8 |
| [16] |
Baucher M, Bernard-Vailhé M A,Chabbert B, et al. Down-regulation of cinnanyl alcohol dehydrogenase in transgenic alfalfa(Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Molecular Biology, 1999,39(3):437-447.
doi: 10.1023/A:1006182925584 |
| [17] |
Li X C, Richard F H . Synthesis and rearrangement reactions of ester-linked lignin-carbohydrate model compounds. Journal of Agricultural and Food Chemistry, 1995,43(8):2098-2103.
doi: 10.1021/jf00056a026 |
| [18] |
Paquette S, Møller B L, Bak S . On the origin of family 1 plant glycosyltransferases. Phytochemistry, 2003,62(3):399-413.
doi: 10.1016/S0031-9422(02)00558-7 |
| [19] |
Lim E K, Bowles D J . A class of plant glycosyltransferases involved in cellular homeostasis. The EMBO Journal, 2004,23(15):2915-2922.
doi: 10.1038/sj.emboj.7600295 |
| [20] |
Bowles D, Isayenkova J, Lim E K , et al. Glycosyltransferases:Managers of small molecules. Current Opinion in Plant Biology, 2005,8(3):254-263.
doi: 10.1016/j.pbi.2005.03.007 |
| [21] |
Gachon C M, Langlois-Meurinne M, Saindrenan P . Plant secondary metabolism glycosyltransferases:The emerging functional analysis. Trends in Plant Science, 2005,10(11):542-549.
doi: 10.1016/j.tplants.2005.09.007 |
| [22] | Lim E K . Plant glycosyltransferases:Their potential as novel biocatalysts. Chemistry:A European Journal, 2005,11(19):5487-5494. |
| [23] |
Li Y, Baldauf S, Lim E K , et al. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. The Journal of Biological Chemistry, 2001,276(6):4338-4343.
doi: 10.1074/jbc.M007447200 |
| [24] | Ross J, Li Y, Lim E , et al. Higher plant glycosyltransferases. Genome Biology, 2001,2(2):31-36. |
| [1] | Wu Ruixiang, Yang Jianchun, Wang Liqin, Guo Xiujuan. Evaluation of the Adaptability of Flax Drought#br# Resistance Based on Multiple Statistics Analysis [J]. Crops, 2018, 34(5): 10-16. |
| [2] | Haibin Luo, Shengli Jiang, Chengmei Huang, Huiqing Cao, Zhinian Deng, Kaichao Wu, Lin Xu, Zhen Lu, Yuanwen Wei. Cloning and Expression of ScHAK10 Gene in Sugarcane [J]. Crops, 2018, 34(4): 53-61. |
| [3] | Xiujuan Guo,Xuejin Feng,Jianchun Yang,Ruixiang Wu,Liqin Wang. Effects of Interaction between Fertilizer and Density on the Yield and Economic Traits of Flax [J]. Crops, 2017, 33(2): 135-138. |
| [4] | Shuyan Wang,Bing Han,Simin Zhou,Jun Xu. Correlation Analysis between the Expression of Stearoyl Acyl Carrier Protein Dehydrogenase Gene and the Soluble Sugar and Fat Content of Oil Flax [J]. Crops, 2016, 32(4): 56-61. |
| [5] | Zanping Han,Yanhui Chen,Shulei Guo,Xiaofeng Zu,Shunxi Wang,Xiyong Zhao. Cloning and Function Analysis of Resistance Gene ZmqLTG3-1 in Maize [J]. Crops, 2016, 32(4): 47-55. |
| [6] | Yu Li,Wenyin Zheng,Chun Feng,Rongfu Wang,Juan Li. Analysis of FeSOD Gene Expression in Normal Wheat Yannong 19 Seedlings under Abiotic Stress [J]. Crops, 2016, 32(4): 75-79. |
| [7] | Yubo Yao,Guangwen Wu,Wengong Huang,Qinghua Kang,Weidong Jiang,Ying Lu,Shuquan Zhang. Potassium Use Efficiency of Different Flax Genotypes [J]. Crops, 2016, 32(4): 80-85. |
| [8] | Xiuxia Cao,Aiping Qian,Wei Zhang,Chongqing Yang. The Effects of Different Amounts of Zinc Fertilizer on the Growth and Yield of Oil-Flax in Arid Land [J]. Crops, 2016, 32(3): 167-170. |
| [9] | Xiujuan Guo,Jianchun Yang,Xuejin Feng,Chao Jiang. Effects of Different Previous Crops in Rotation System on Dry Matter Accumulation,Quality,and Yield in Flax [J]. Crops, 2016, 32(2): 164-167. |
| [10] | Li Zhao,Limin Wang,Wei Zhao,Zhao Dang,Jianping Zhang,Zhanhai Dang. The Isolation of Crude Fat and Fatty Acid Components in a RIL Population of Oil Flax and Their Correlation Analysis [J]. Crops, 2016, 32(1): 33-37. |
|
||