Crops ›› 2017, Vol. 33 ›› Issue (6): 37-44.doi: 10.16035/j.issn.1001-7283.2017.06.007
Previous Articles Next Articles
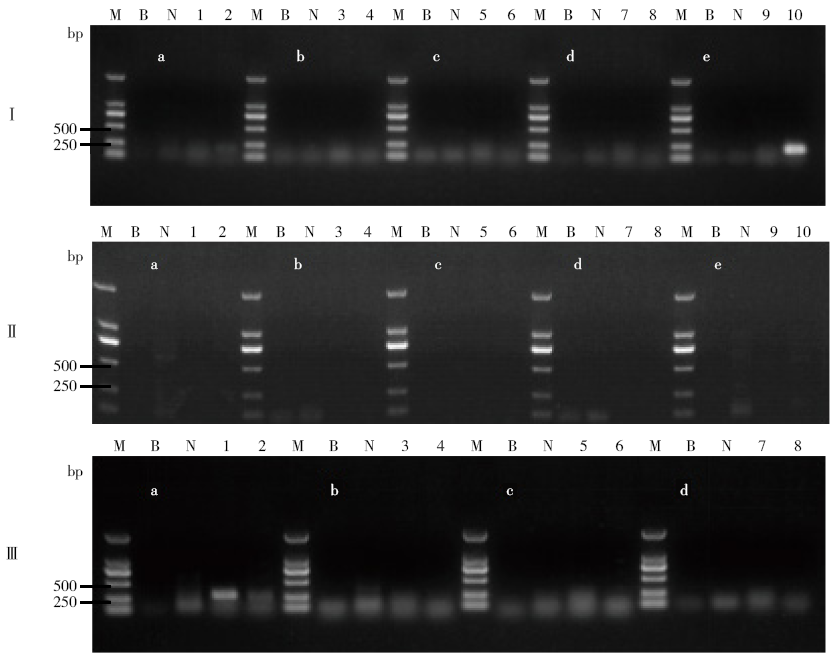
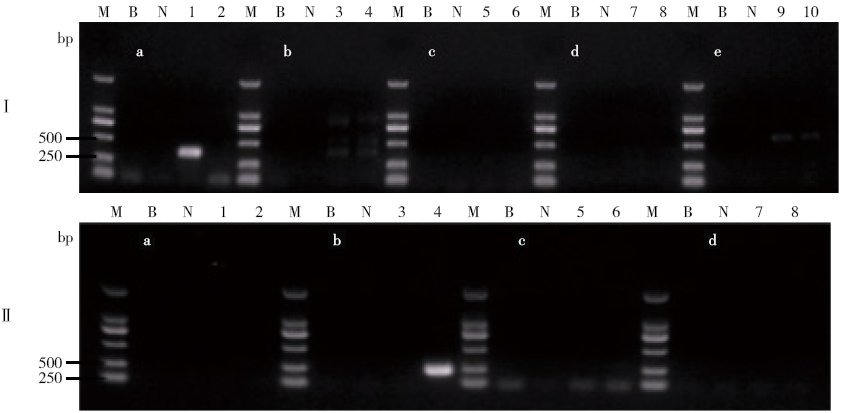
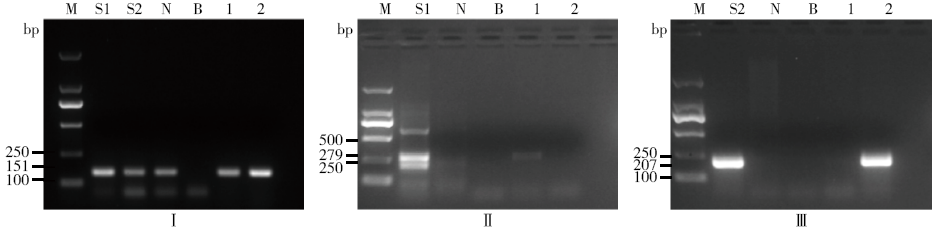
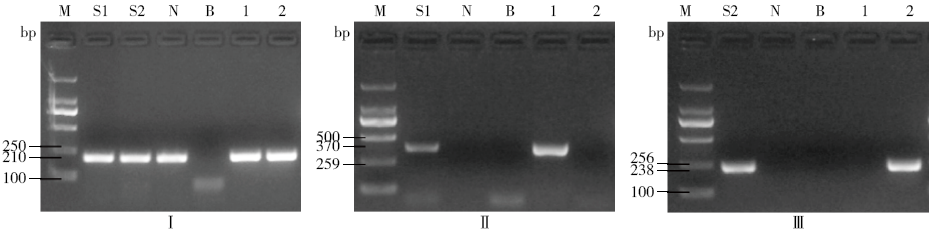
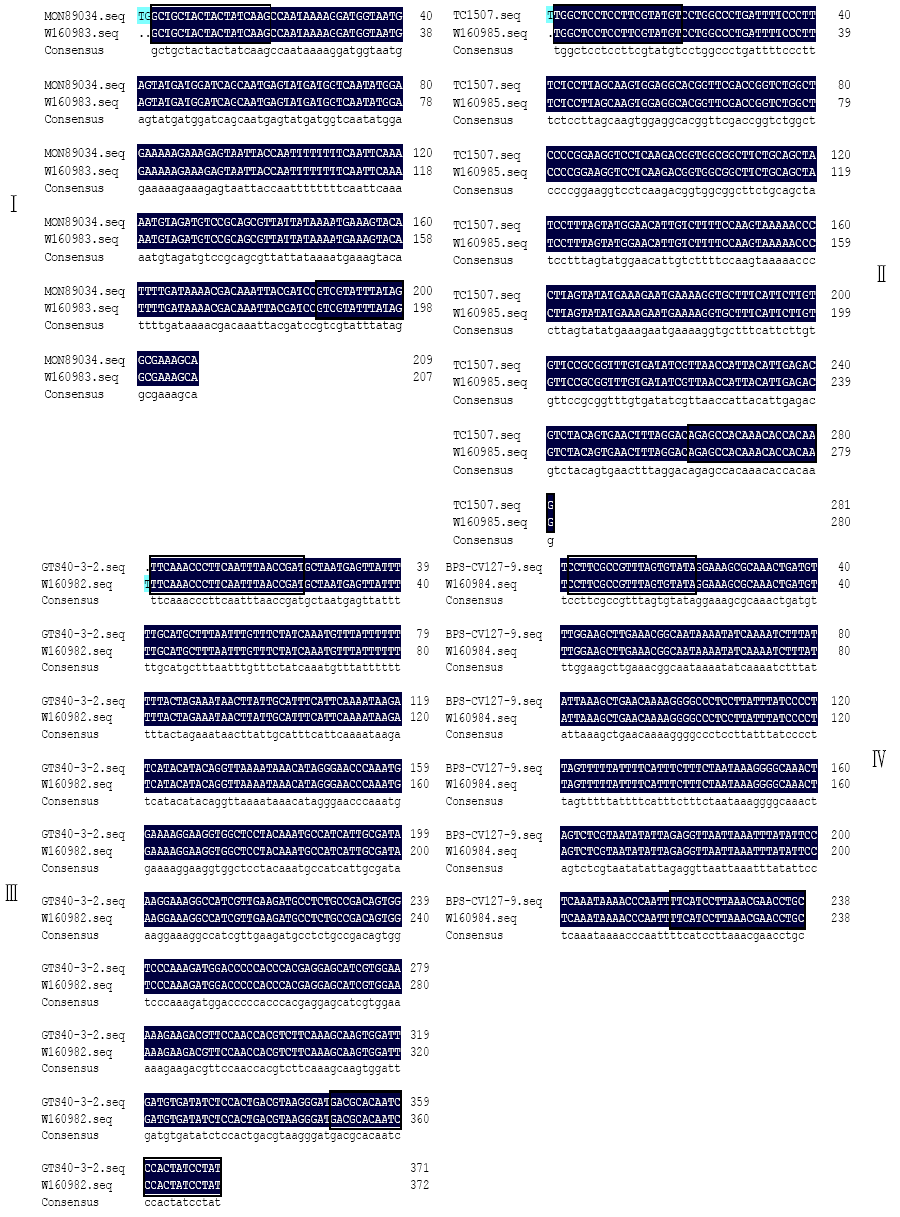
Determining Blind Samples of Transgenic Maize and Transgenic Soybean
Li Lina,Jin Longguo,Xie Chuanxiao,Liu Changlin
- Institute of Crop Sciences,Chinese Academy of Agricultural Sciences,Beijing 100081,China
| [1] | James C . 2016年全球生物技术/转基因作物商业化发展态势. 中国生物工程杂志, 2017,37(4):1-8. |
| [2] | James C . 2015年全球生物技术/转基因作物商业化发展态势. 中国生物工程杂志, 2016,36(4):1-11. |
| [3] | 姜天海 . 全球转基因种植面积再创历史新高.科学新闻, 2017(5):75-77. |
| [4] |
沈平, 章秋艳, 林友华 , 等. 推进我国转基因玉米产业化的思考. 中国生物工程杂志, 2016,36(4):24-29.
doi: 10.13523/j.cb.20160404 |
| [5] | 李楠 . 转基因植物基因漂移及生态风险研究的文献计量学分析. 江苏农业科学, 2015,43(12):62-67. |
| [6] | 郭建英, 万方浩, 韩召军 . 转基因植物的生态安全性风险. 中国生态农业学报, 2008,16(2):515-522. |
| [7] | 付仲文 . 中国农业转基因生物安全管理. 转基因生物与环境国际研讨会, 2004. |
| [8] |
熊建文, 彭端, 韦剑锋 . 转基因玉米研究与应用进展. 广东农业科学, 2012,39(6):27-29.
doi: 10.3969/j.issn.1004-874X.2012.06.010 |
| [9] | GB/T 1861-3-2012 B/T 1861-3-2012.转基因植物及其产品成分检测玉米内标准基因定性PCR方法.北京: 中国标准出版社, 2012. |
| [10] | GB/T 1485-15-2010 B/T 1485-15-2010.转基因植物及其产品成分检测抗虫耐除草剂玉米MON88017及其衍生品种定性PCR方法.北京: 中国标准出版社, 2010. |
| [11] | GB/T 869-12-2007 B/T 869-12-2007.转基因植物及其产品成分检测耐除草剂玉米GA21及其衍生品种定性PCR方法.北京: 中国标准出版社, 2007. |
| [12] | GB/T 1485-16-2010 B/T 1485-16-2010.转基因植物及其产品成分检测抗虫玉米MIR604及其衍生品种定性PCR方法.北京: 中国标准出版社, 2010. |
| [13] | GB/T 2031-5-2013 B/T 2031-5-2013.转基因植物及其产品成分检测耐旱玉米MON87460及其衍生品种定性PCR方法.北京: 中国标准出版社, 2013. |
| [14] | GB/T 1861-4-2012 B/T 1861-4-2012.转基因植物及其产品成分检测抗虫玉米MON89034及其衍生品种定性PCR方法.北京: 中国标准出版社, 2012. |
| [15] | GB/T 2031-6-2013 B/T 2031-6-2013.转基因植物及其产品成分检测抗虫玉米MIR162及其衍生品种定性PCR方法.北京: 中国标准出版社, 2013. |
| [16] | GB/T 2122-16-2014 B/T 2122-16-2014.转基因植物及其产品成分检测抗虫玉米MON810及其衍生品种定性PCR方法.北京: 中国标准出版社, 2014. |
| [17] | GB/T 869-10-2007 B/T 869-10-2007.转基因植物及其产品成分检测抗虫玉米MON863及其衍生品种定性PCR方法.北京: 中国标准出版社, 2007. |
| [18] | GB/T 2122-15-2014 B/T 2122-15-2014.转基因植物及其产品成分检测抗虫和耐除草剂玉米Bt176及其衍生品种定性PCR方法.北京: 中国标准出版社, 2014. |
| [19] | GB/T 2122-9-2014 B/T 2122-9-2014.转基因植物及其产品成分检测耐除草剂玉米DAS-40278-9及其衍生品种定性PCR方法.北京: 中国标准出版社, 2014. |
| [20] | GB/T 869-7-2007 869-7-2007.转基因植物及其产品成分检测抗虫和耐除草剂玉米TC1507及其衍生品种定性PCR方法.北京: 中国标准出版社, 2007. |
| [21] | GB/T 2031-13-2013 B/T 2031-13-2013.转基因植物及其产品成分检测转淀粉酶基因玉米3272及其衍生品种定性PCR方法.北京: 中国标准出版社, 2013. |
| [22] | GB/T 1485-9-2010 B/T 1485-9-2010.转基因植物及其产品成分检测抗虫耐除草剂玉米59122及其衍生品种定性PCR方法.北京: 中国标准出版社, 2010. |
| [23] | GB/T 869-13-2007 B/T 869-13-2007.转基因植物及其产品成分检测耐除草剂玉米NK603及其衍生品种定性PCR方法.北京: 中国标准出版社, 2007. |
| [24] | GB/T 2031-8-2013 B/T 2031-8-2013.转基因植物及其产品成分检测大豆内标准基因定性PCR方法.北京: 中国标准出版社, 2013. |
| [25] | GB/T 1861-2-2012 B/T 1861-2-2012.转基因植物及其产品成分检测耐除草剂大豆GTS 40-3-2及其衍生品种定性PCR方法.北京: 中国标准出版社, 2012. |
| [26] | GB/T 1485-6-2010 B/T 1485-6-2010.转基因植物及其产品成分检测耐除草剂大豆MON89788及其衍生品种定性PCR方法.北京: 中国标准出版社, 2010. |
| [27] | GB/T 2031-8-2013 B/T 2031-8-2013.农业部2259号公告-7-2015转基因植物及其产品成分检测抗虫大豆MON87701及其衍生品种定性PCR方法.北京: 中国标准出版社, 2013. |
| [28] | GB/T 2122-4-2014 B/T 2122-4-2014.转基因植物及其产品成分检测耐除草剂和品质改良大豆MON87705及其衍生品种定性PCR方法.北京: 中国标准出版社, 2014. |
| [29] | GB/T 2259-6-2015 B/T 2259-6-2015.转基因植物及其产品成分检测耐除草剂大豆MON87708及其衍生品种定性PCR方法.北京: 中国标准出版社, 2015. |
| [30] | GB/T 2122-5-2014 B/T 2122-5-2014.转基因植物及其产品成分检测品质改良大豆MON87769及其衍生品种定性PCR方法.北京: 中国标准出版社, 2014. |
| [31] | GB/T 1782-5-2012 B/T 1782-5-2012.转基因植物及其产品成分检测耐除草剂大豆CV127及其衍生品种定性PCR方法.北京: 中国标准出版社, 2012. |
| [32] | GB/T 1782-4-2012 B/T 1782-4-2012.转基因植物及其产品成分检测高油酸大豆305423及其衍生品种定性PCR方法.北京: 中国标准出版社, 2012. |
| [33] | GB/T 1782-1-2012 B/T 1782-1-2012.转基因植物及其产品成分检测耐除草剂大豆356043及其衍生品种定性PCR方法.北京: 中国标准出版社, 2012. |
| [34] |
李荣华, 夏岩石, 刘顺枝 , 等. 改进的CTAB提取植物DNA方法. 实验室研究与探索, 2009,28(9):14-16.
doi: 10.3969/j.issn.1006-7167.2009.09.005 |
| [35] | GB/T 3576-2013 B/T 3576-2013.转基因成分检测大豆PCR-DHPLC检测方法.北京: 中国标准出版社, 2013. |
| [1] | Cui Guo,Wei Zhang,Guirong Yu,Zhengfu Zhou,Liang Li,Shuai Feng,Ming Chen,Jin Wang. Analysis of the Flanking Sequence and Event-Specific Detection of Transgenic G2-EPSPS Line of Maize [J]. Crops, 2016, 32(1): 69-75. |
|