Crops ›› 2018, Vol. 34 ›› Issue (5): 54-62.doi: 10.16035/j.issn.1001-7283.2018.05.009
Previous Articles Next Articles
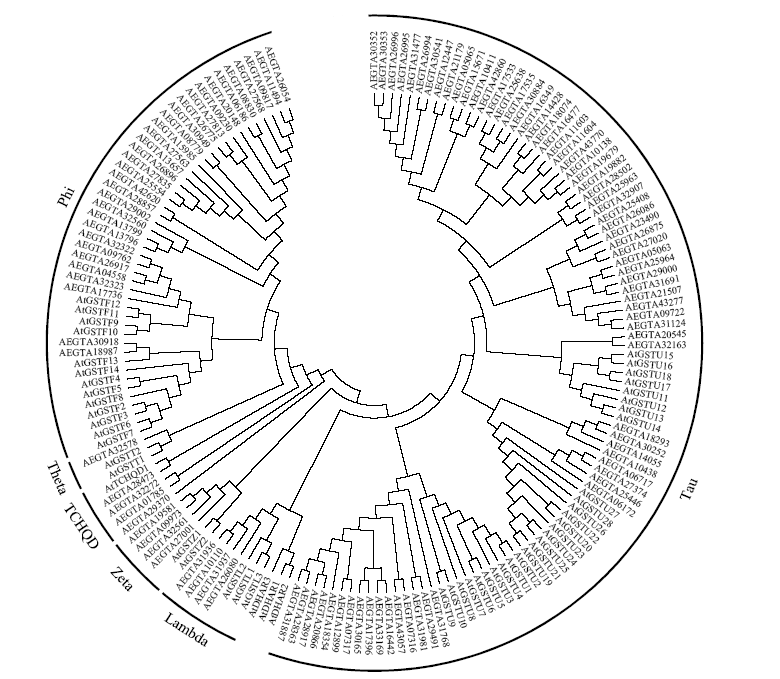
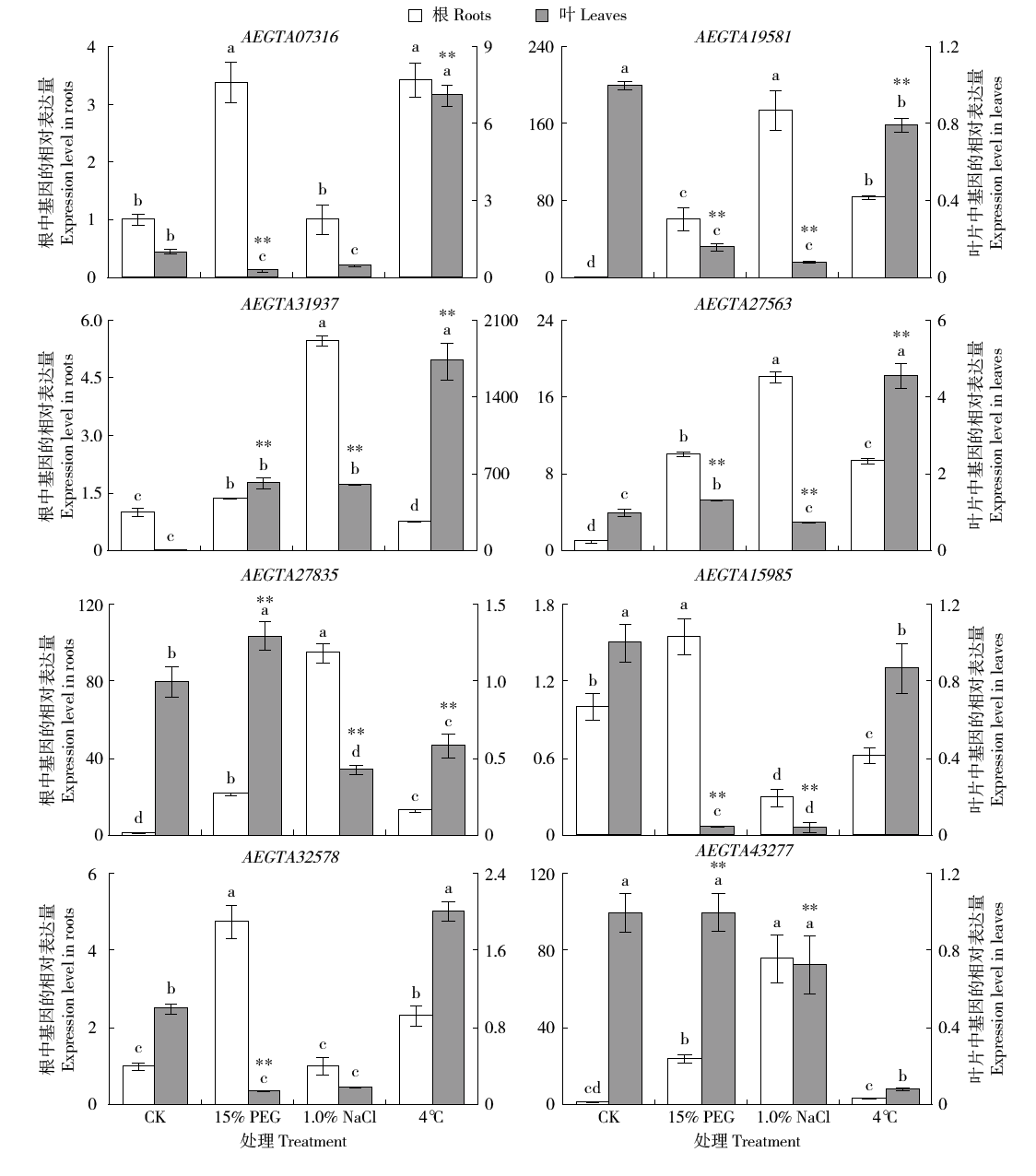
Identification and Expression Analysis of the Whole Glutathione S-Transferase Genome Family in Aegilops tauschii under Abiotic Stress
Ma Jianhui,Zhang Wenli,Gao Xiaolong,Zhang Daijing,Jiang Lina,Zhai Yanyu,Shao Yun,Li Chunxi
- College of Life Sciences, Henan Normal University, Xinxiang 453007, Henan, China
| [1] | Sies H . Glutathione and its role in cellular functions. Free Radical Biology & Medicine, 1999,27(9):916-921. |
| [2] |
Edwards R, Dixon D P, Walbot V . Plant glutathione S-transferases:enzymes with multiple functions in sickness and in health. Trends in Plant Science, 2000,5(5):193-198.
doi: 10.1016/S1360-1385(00)01601-0 |
| [3] |
Öztetik E . A tale of plant glutathione S-transferases:Since 1970. Botanical Review, 2008,74(3):419-437.
doi: 10.1007/s12229-008-9013-9 |
| [4] |
Cummins I, Dixon D P, Freitag-Pohl S , et al. Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metabolism Reviews, 2011,43(2):266-280.
doi: 10.3109/03602532.2011.552910 |
| [5] | Marrs K A . The functions and regulation of glutathione S-transferases in plants. Annual Review of Plant Physiology & Plant Molecular Biology, 1996,47(1):127-158. |
| [6] |
Sheehan D, Meade G, Foley V M . Structure,function and evolution of glutathione transferases:implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochemical Journal, 2001,360(1):1-16.
doi: 10.1042/bj3600001 |
| [7] | Dixon D P, Lapthorn A, Edwards R . Plant glutathione transferases. Genome Biology, 2002,401(3):169-186. |
| [8] |
McGonigle B, Keeler S J, Lau S M C , et al. A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiology, 2000,124(3):1105-1120.
doi: 10.1104/pp.124.3.1105 |
| [9] |
Soranzo N, Gorla M S, Mizzi L , et al. Organisation and structural evolution of the rice glutathione S-transferase gene family. Molecular Genetics and Genomics, 2004,271(5):511-521.
doi: 10.1007/s00438-004-1006-8 |
| [10] | 江董丽, 才华, 柏锡 , 等. 大豆GST基因家族全基因组筛选、分类和表达. 分子植物育种, 2013,4(5):465-475. |
| [11] | 李晓玉, 江海波, 江海洋 , 等. 玉米全基因组谷胱苷肽-S-转移酶基因家族的分析. 安徽农业大学学报, 2013,40(3):350-356. |
| [12] |
Wu J, Cramer C L, Hatzios K K . Characterization of two cDNAs encoding glutathione S-transferases in rice and induction of their transcripts by the herbicide safener fenclorim. Physiologia Plantarum, 1999,105(1):102-108.
doi: 10.1034/j.1399-3054.1999.105116.x |
| [13] |
Moons A . Osgstu3 and Osgtu4,encoding tau class glutathione S-transferases,are heavy metal and hypoxic stress-induced and differentially salt stress-responsive in rice roots. FEBS Letters, 2003,553(3):427-432.
doi: 10.1016/S0014-5793(03)01077-9 |
| [14] | 李永生, 方永丰, 李玥 , 等. 玉米逆境响应基因ZmGST23克隆和表达分析. 农业生物技术学报, 2016,24(5):667-677. |
| [15] | Wang Z Y, Cai H, Bai X , et al. Isolation of GsGST19 from Glycine soja and analysis of Saline-Alkaline tolerance for transgenic Medicago sativa. Acta Agronomica Sinica, 2012,38(6):971-979. |
| [16] | 韩少怀, 李佳佳, 张璟曜 , 等. 大豆GmGSTl2基因的克隆及表达分析. 大豆科学, 2015(5):782-788. |
| [17] |
Rezaei M K, Shobbar Z S, Shahbazi M , et al. Glutathione S-transferase (GST) family in barley:identification of members,enzyme activity,and gene expression pattern. Journal of Plant Physiology, 2013,170(14):1277-1284.
doi: 10.1016/j.jplph.2013.04.005 |
| [18] |
Williamson G, Beverley M C . Wheat glutathione S-transferase:purification and properties. Journal of Cereal Science, 1988,8(2):155-163.
doi: 10.1016/S0733-5210(88)80026-2 |
| [19] |
Mauch F, Hertig C, Rebmann G , et al. A wheat glutathione-S-transferase gene with transposon-like sequences in the promoter region. Plant Molecular Biology, 1991,16(6):1089-1091.
doi: 10.1007/BF00016083 |
| [20] | 吴金华, 张西平, 胡言光 , 等. 小麦抗白粉病相关基因GST克隆与表达. 西北植物学报, 2013(1):34-38. |
| [21] |
Dixon D, Cole D J, Edwards R . Characterisation of multiple glutathione transferases containing the GST I subunit with activities toward herbicide substrates in maize (Zea mays). Pest Management Science, 1997,50(1):72-82.
doi: 10.1002/(SICI)1096-9063(199705)50:1<>1.0.CO;2-C |
| [22] |
Huala E, Dickerman A W, Garcia-Hernandez M , et al. The Arabidopsis information resource (TAIR):a comprehensive database and web-based information retrieval,analysis,and visualization system for a model plant. Nucleic Acids Research, 2001,29(1):102-105.
doi: 10.1093/nar/29.1.102 |
| [23] |
Finn R D, Mistry J, Schuster-Böckler B , et al. Pfam:clans,web tools and services. Nucleic Acids Research, 2006,34(s1):247-251.
doi: 10.1093/nar/gkj149 |
| [24] | Letunic I, Doerks T, Bork P . SMART 6:recent updates and new developments. Nucleic Acids Research, 2009,37(s1):229-232. |
| [25] |
Suyama M, Torrents D, Bork P . PAL2NAL:robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Research, 2006,34(s2):609-612.
doi: 10.1093/nar/gkl315 |
| [26] |
Gaut B S, Doebley J F . DNA sequence evidence for the segmental allotetraploid origin of maize. Proceedings of the National Academy of Sciences of the United States of America, 1997,94(13):6809-6814.
doi: 10.1073/pnas.94.13.6809 |
| [27] |
Peng X, Zhao Y, Cao J , et al. CCCH-type zinc finger family in maize:genome-wide identification,classification and expression profiling under abscisic acid and drought treatments. PloS One, 2012,7(7):e40120.
doi: 10.1371/journal.pone.0040120 |
| [28] |
Frova C . The plant glutathione transferase gene family:genomic structure,functions,expression and evolution. Physiologia Plantarum, 2003,119(4):469-479.
doi: 10.1046/j.1399-3054.2003.00183.x |
| [29] | 张雪, 陶磊, 乔晟 , 等. 谷胱甘肽转移酶在植物抵抗非生物胁迫方面的角色. 中国生物工程杂志, 2017,37(3):92-98. |
| [30] |
Wagner U, Edwards R, Dixon D P , et al. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Molecular Biology, 2002,49(5):515-532.
doi: 10.1023/A:1015557300450 |
| [31] | 戚元成, 张世敏, 王丽萍 , 等. 谷胱甘肽转移酶基因过量表达能加速盐胁迫下转基因拟南芥的生长. 植物生理与分子生物学学报, 2004,30(5):517-522. |
| [32] |
Liu D, Liu Y, Rao J , et al. Overexpression of the glutathione S-transferase gene from Pyrus pyrifolia fruit improves tolerance to abiotic stress in transgenic tobacco plants. Molecular Biology, 2013,47(4):515-523.
doi: 10.1134/S0026893313040109 |
| [33] |
Ji W, Zhu Y, Li Y , et al. Over-expression of a glutathione S-transferase gene,GsGST,from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotechnology Letters, 2010,32(8):1173-1179.
doi: 10.1007/s10529-010-0269-x |
| [34] |
George S, Venkataraman G, Parida A . A chloroplast-localized and auxin-induced glutathione S-transferase from phreatophyte Prosopis juliflora confer drought tolerance on tobacco. Journal of Plant Physiology, 2010,167(4):311-318.
doi: 10.1016/j.jplph.2009.09.004 |
| [35] |
Xu J, Xing X J, Tian Y S , et al. Transgenic Arabidopsis plants expressing tomato glutathione S-transferase showed enhanced resistance to salt and drought stress. PloS One, 2015,10(9):e0136960.
doi: 10.1371/journal.pone.0136960 |
| [36] |
Yang G, Xu Z, Peng S , et al. In planta characterization of a tau class glutathione S-transferase gene from Juglans regia (JrGSTTau1) involved in chilling tolerance. Plant Cell Reports, 2016,35(3):681-692.
doi: 10.1007/s00299-015-1912-8 |
| [37] |
Zhao J, Zhang S, Yang T , et al. Global transcriptional profiling of a cold-tolerant rice variety under moderate cold stress reveals different cold stress response mechanisms. Physiologia Plantarum, 2015,154(3):381-394.
doi: 10.1111/ppl.2015.154.issue-3 |
| [38] |
Le Martret B, Poage M, Shiel K , et al. Tobacco chloroplast transformants expressing genes encoding dehydroascorbate reductase,glutathione reductase,and glutathione-S-transferase,exhibit altered anti-oxidant metabolism and improved abiotic stress tolerance. Plant Biotechnology Journal, 2011,9(6):661-673.
doi: 10.1111/pbi.2011.9.issue-6 |
|