Crops ›› 2020, Vol. 36 ›› Issue (5): 41-47.doi: 10.16035/j.issn.1001-7283.2020.05.006
Previous Articles Next Articles
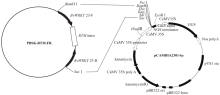
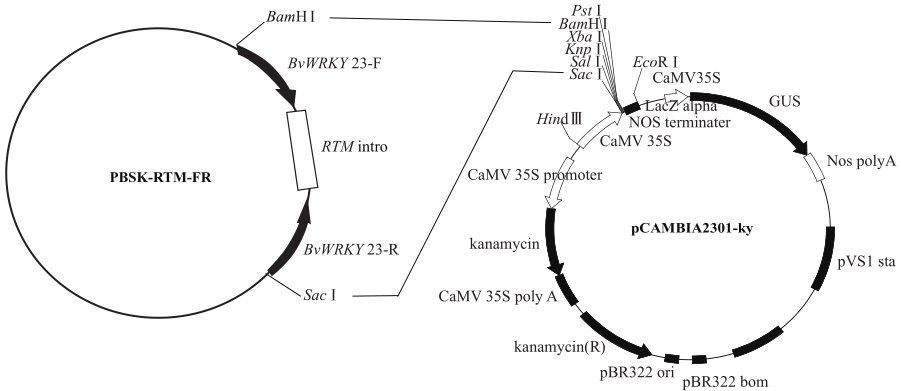
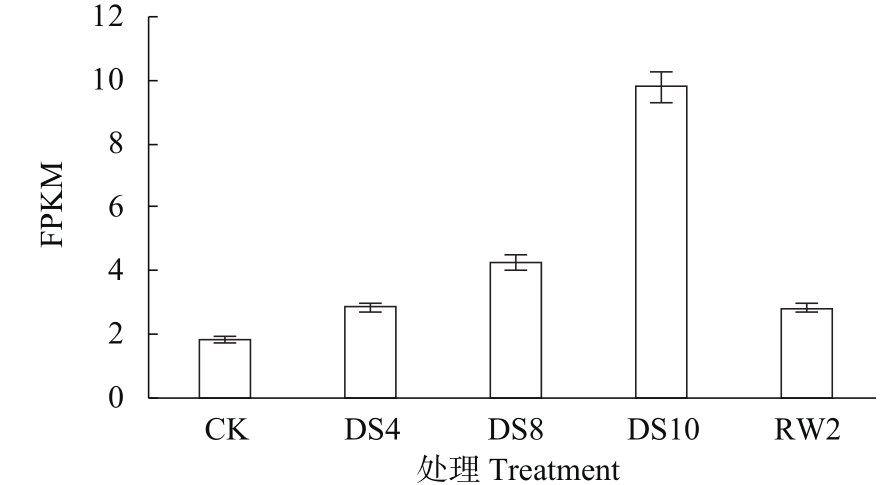
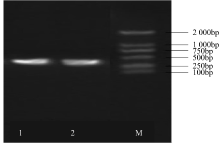
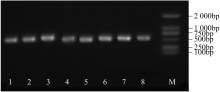
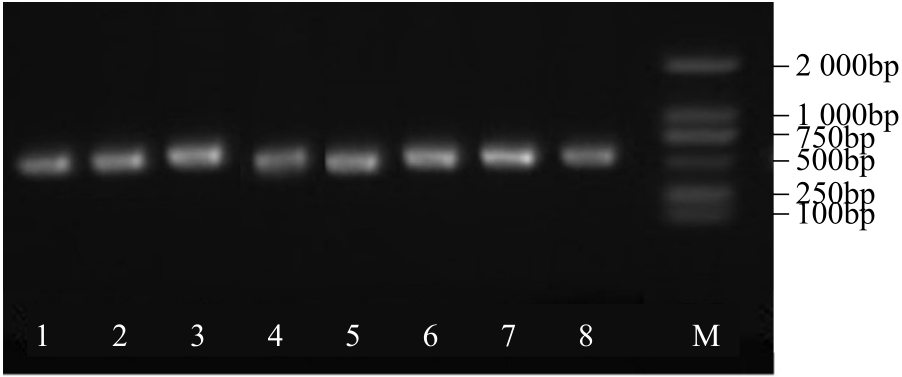
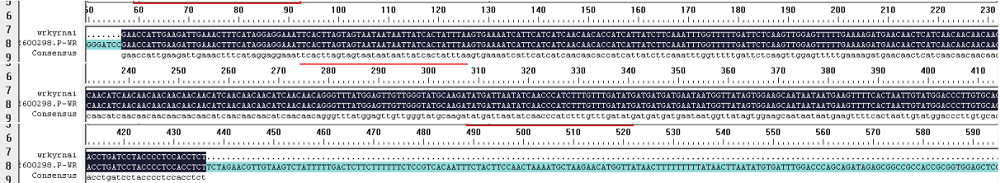
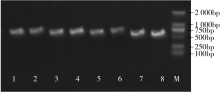
Construction of RNAi Expression Vector of BvWRKY23 Gene in Sugar Beet
Li Guolong1( ), Wu Haixia2, Sun Yaqing1
), Wu Haixia2, Sun Yaqing1
- 1College of Agronomy, Inner Mongolia Agricultural University, Hohhot 010018, Inner Mongolia, China
2Inner Mongolia Research Institute for Water Conservancy, Hohhot 010020, Inner Mongolia, China
| [1] | Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein,SPF1,that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Molecular Genetics and Genomics, 1994,244(6):563-571. |
| [2] |
Chen W Q, Provart N J, Glazebrook J, et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. The Plant Cell, 2002,14(3):559-574.
doi: 10.1105/tpc.010410 pmid: 11910004 |
| [3] |
Xu X P, Chen C H, Fan B F, et al. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18,WRKY40,and WRKY60 transcription factors. The Plant Cell, 2006,18(5):1310-1326.
doi: 10.1105/tpc.105.037523 pmid: 16603654 |
| [4] |
Ramamoorthy R, Jiang S Y, Kumar N, et al. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant and Cell Physiology, 2008,49(6):865-879.
doi: 10.1093/pcp/pcn061 pmid: 18413358 |
| [5] |
Christian R, Shen Q X. The WRKY gene family in rice (Oryza sativa). Journal of Integrative Plant Biology, 2007,49(6):827-842.
doi: 10.1111/j.1744-7909.2007.00504.x |
| [6] |
Ning P, Liu C C, Kang J Q, et al. Genome-wide analysis of WRKY transcription factors in wheat (Triticum aestivum L.) and differential expression under water deficit condition. PeerJ, 2017,5:e3232.
doi: 10.7717/peerj.3232 pmid: 28484671 |
| [7] |
Mangelsen E, Kilian J, Berendzen K W, et al. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics, 2008,9(1):194.
doi: 10.1186/1471-2164-9-194 |
| [8] |
Wei K F, Chen J, Chen Y F, et al. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Research, 2012,19(2):153-164.
doi: 10.1093/dnares/dsr048 |
| [9] |
Yang Y, Zhou Y, Chi Y, et al. Characterization of soybean WRKY gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode. Scientific Reports, 2017,7(1):17804.
doi: 10.1038/s41598-017-18235-8 pmid: 29259331 |
| [10] |
Dellagi A, Birch P R J, Heilbronn J, et al. A potato gene,erg-1,is rapidly induced by Erwinia carotovora ssp. atroseptica,Phytophthora infestans,ethylene and salicylic acid. Journal of Plant Physiology, 2000,157(2):201-205.
doi: 10.1016/S0176-1617(00)80191-1 |
| [11] |
祖倩丽, 尹丽娟, 徐兆师, 等. 谷子WRKY36转录因子的分子特性及功能鉴定. 中国农业科学, 2015,48(5):851-860.
doi: 10.3864/j.issn.0578-1752.2015.05.03 |
| [12] |
孔维龙, 于坤, 但乃震, 等. 甜菜WRKY转录因子全基因组鉴定及其在非生物胁迫下的表达分析. 中国农业科学, 2017,50(17):3259-3273.
doi: 10.3864/j.issn.0578-1752.2017.17.002 |
| [13] |
Yao D X, Zhang X Y, Zhao X H, et al. Transcriptome analysis reveals salt-stress-regulated biological processes and key pathways in roots of cotton (Gossypium hirsutum L.). Genomics, 2011,98(1):47-55.
doi: 10.1016/j.ygeno.2011.04.007 |
| [14] |
Ülker B, Somssich I E. WRKY transcription factors:from DNA binding towards biological function. Current Opinion in Plant Biology, 2004,7(5):491-498.
doi: 10.1016/j.pbi.2004.07.012 |
| [15] |
Rushton P J, Somssich I E, Ringler P, et al. WRKY transcription factors. Trends in Plant Science, 2010,15(5):247-258.
doi: 10.1016/j.tplants.2010.02.006 |
| [16] |
Jiang Y J, Liang G, Yu D Q. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Molecular Plant, 2012,5(6):1375-1388.
doi: 10.1093/mp/sss080 |
| [17] |
Chen H, Lai Z R, Shi J W, et al. Roles of Arabidopsis WRKY18,WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biology, 2010,10:281.
doi: 10.1186/1471-2229-10-281 pmid: 21167067 |
| [18] |
Pandey S P, Roccaro M, Schon M, et al. Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. The Plant Journal, 2010,64(6):912-923.
doi: 10.1111/j.1365-313X.2010.04387.x pmid: 21143673 |
| [19] |
Schon M, Toller A, Diezel C, et al. Analyses of wrky18 wrky40 plants reveal critical roles of SA/EDS1 signaling and indole-glucosinolate biosynthesis for Golovinomyces orontii resistance and a loss-of resistance towards Pseudomonas syringae pv. tomato AvrRPS4. Molecular Plant-Microbe Interactions, 2013,26(7):758-767.
doi: 10.1094/MPMI-11-12-0265-R pmid: 23617415 |
| [20] |
Babitha K C, Ramu S V, Pruthvi V, et al. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Research, 2013,22(2):327-341.
doi: 10.1007/s11248-012-9645-8 |
| [21] |
Wu X L, Shiroto Y, Kishitani S, et al. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Reports, 2009,28(1):21-30.
doi: 10.1007/s00299-008-0614-x |
| [22] |
Tao Z, Kou Y J, Liu H B, et al. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. Journal of Experimental Botany, 2011,62(14):4863-4874.
doi: 10.1093/jxb/err144 |
| [23] |
Shen H S, Liu C T, Zhang Y, et al. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Molecular Biology, 2012,80(3):241-253.
doi: 10.1007/s11103-012-9941-y |
| [24] |
Raineri J, Wang S H, Peleg Z, et al. The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Molecular Biology, 2015,88(4):401-413.
doi: 10.1007/s11103-015-0329-7 |
| [25] |
Lee H, Cha J, Choi C, et al. Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice, 2018,11(1):5.
doi: 10.1186/s12284-018-0199-0 pmid: 29330772 |
| [26] |
Gao H M, Wang Y F, Xu P, et al. Overexpression of a WRKY transcription factor Tawrky2 enhances drought stress tolerance in transgenic wheat. Frontiers in Plant Science, 2018,9:997.
doi: 10.3389/fpls.2018.00997 pmid: 30131813 |
| [27] |
Wang C T, Ru J N, Liu Y W, et al. Maize WRKY transcription factor Zmwrky106 confers drought and heat tolerance in transgenic plants. International Journal of Molecular Sciences, 2018,19(10):3046.
doi: 10.3390/ijms19103046 |
| [28] | 李大红, 王春弘, 刘喜平, 等. 大豆GmWRKY35基因的克隆及其增强烟草耐旱能力研究. 大豆科学, 2017,36(5):685-691. |
| [29] | 韩凯虹, 张继宗, 王伟婧, 等. 水分胁迫及复水对华北寒旱区甜菜生长及品质的影响. 灌溉排水学报, 2015,34(4):63-68. |
| [30] | 李国龙, 吴海霞, 温丽, 等. 甜菜苗期抗旱鉴定指标筛选及其综合评价. 干旱地区农业研究, 2011,29(4):69-74. |
| [31] | 王伟伟, 刘妮, 陆沁, 等. RNAi技术的最新研究进展. 生物技术通报, 2017,33(11):35-40. |
| [32] |
Fire A, Xu S, Montgomery M K, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 1998,391(6669):806-811.
doi: 10.1038/35888 pmid: 9486653 |
| [33] |
Wang L Q, Li Z, Lu M Z, et al. ThNAC13,a NAC transcription factor from Tamarix hispida,confers salt and osmotic stress tolerance to transgenic Tamarix and Arabidopsis. Frontiers in Plant Science, 2017,8:635.
doi: 10.3389/fpls.2017.00635 pmid: 28491072 |
| [34] |
Xu H J, Xue J, Lu B, et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature, 2015,519(7544):464-467.
doi: 10.1038/nature14286 pmid: 25799997 |
| [35] |
Tang B, Wei P, Zhao L N, et al. Knockdown of five trehalase genes using RNA interference regulates the gene expression of the chitin biosynthesis pathway in Tribolium castaneum. BMC Biotechnology, 2016,16:67.
doi: 10.1186/s12896-016-0297-2 pmid: 27596613 |
| [36] |
Hu Z, Lin Q, Chen H, et al. Identification of a novel cytochrome P450 gene,CYP321E1 from the diamondback moth,Plutella xylostella (L.) and RNA interference to evaluate its role in chlorantraniliprole resistance. Bulletin of Entomological Research, 2014,104(6):716-723.
doi: 10.1017/S0007485314000510 |
| [37] |
Guo Z J, Kang S, Zhu X, et al. The novel ABC transporter ABCH1 isa potential target for RNAi-based insect pest control and resistance management. Scientific Reports, 2015,5:13728.
doi: 10.1038/srep13728 pmid: 26333918 |
| [38] |
Li L, Li Q Y, Bao Y H, et al. RNAi-based inhibition of porcine reproductive and respiratorysyndrome virus replication intransgenic pigs. Journal of Biotechnology, 2014,171:17-24.
doi: 10.1016/j.jbiotec.2013.11.022 |
| [39] |
Linke L M, Wilusz J, Pabilonia K L, et al. Inhibiting avian influenza virus shedding using a novel RNAi antiviral vector technology:proof of concept inan avian cell model. AMB Express, 2016,6(1):16.
doi: 10.1186/s13568-016-0187-y pmid: 26910902 |
| [40] |
Wu S H, Wen F F, Li Y Y, et al. PIK3CA and PIK3CB silencing by RNAi reverse MDR and inhibit tumorigenic properties in human colorectal carcinoma. Tumor Biology, 2016,37:8799-8809.
doi: 10.1007/s13277-015-4691-5 pmid: 26747178 |
| [41] |
Wang F J, Chen W C, Liu P F, et al. Lentivirus-mediated RNAi knockdown of LMP2A inhibits the growth of the Epstein-Barrassociated gastric carcinoma cell line GT38 in vitro. Experimental and Therapeutic Medicine, 2017,13:187-193.
doi: 10.3892/etm.2016.3954 pmid: 28123488 |
| [42] | Weinsteina S, Toker I A, Emmanuel R, et al. Harnessing RNAi-based nanomedicines for therapeutic gene silencing in B-cell malignancies. Proceedings of the National Academy Sciences of the United States of America, 2015,113(1):E16-E22. |
| [43] | 柴晓杰, 王丕武, 关淑艳, 等. 玉米淀粉分支酶基因反义表达载体的构建和功能分析. 作物学报, 2005,31(12):1654-1656. |
| [44] | Chen S B, Pattavipha S, Liu J L, et al. A versatile zero background T-vector system for gene cloning and functional genomics1 [C][W][OA]. Plant Physiology, 2009,150(3):1111-1121. |
| [45] | 姜玲, 秦长平, 伏卉. Gateway TM系统快速构建番木瓜环斑病毒CP基因反向重复序列表达载体 . 农业生物技术学报, 2008,16(3):526-529. |
| [46] |
Schwab R, Ossowski S, Riester M, et al. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell, 2006,18:1121-1133.
doi: 10.1105/tpc.105.039834 pmid: 16531494 |
| [47] | 邢珍娟, 王振营, 何康来, 等. 转Bt基因玉米幼苗残体中CrylAb杀虫蛋白田间降解动态. 中国农业科学, 2008,41(2):412-416. |
| [48] | 王镭, 才华, 柏锡, 等. 转OsCDPK7基因水稻的培育与耐盐性分析. 遗传, 2008,30(8):1051-1052. |
| [49] | 吕品, 柴晓杰, 王丕武, 等. 大豆胰蛋白酶抑制剂KSTI3基因的克隆及其植物表达载体的构建. 吉林农业大学学报, 2007,29(3):275-278. |
| [50] | 娄玲玲, 郑唐春, 曲冠证, 等. 可用于植物转化的RNAi载体构建方法. 安徽农业大学学报, 2012,39(1):120-123. |
| [51] |
Manamohan M, Sharath Chandra G, Asokan R, et al. One-step DNA fragment assembly for expressing intron-containing hairpin RNA in plants for gene silencing. Analytical Biochemistry, 2013,433(2):189-191.
doi: 10.1016/j.ab.2012.09.034 |
|
||