Crops ›› 2023, Vol. 39 ›› Issue (5): 43-48.doi: 10.16035/j.issn.1001-7283.2023.05.007
Previous Articles Next Articles
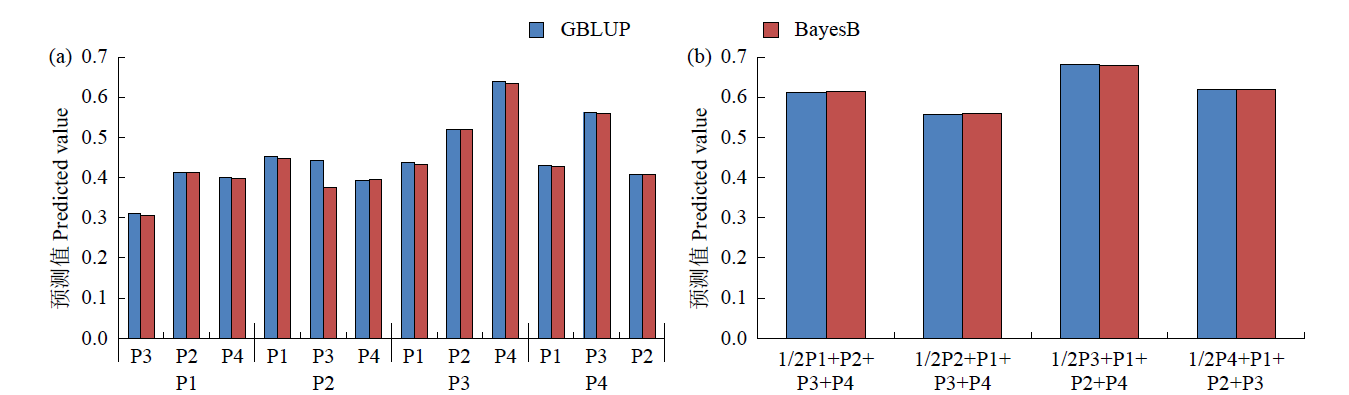
Whole-Genome Predictive Analysis of Fresh Weight per Plant Using the Maize F1 Population
Yang Zongying1( ), Xiao Gui2(
), Xiao Gui2( ), Zhang Hongwei3(
), Zhang Hongwei3( )
)
- 1College of Agronomy, Jilin Agricultural University, Changchun 130118, Jilin, China
2Dingxi Academy of Agricultural Sciences, Dingxi 743000, Gansu, China
3Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100193, China
| [1] |
李少昆, 赵久然, 董树亭, 等. 中国玉米栽培研究进展与展望. 中国农业科学, 2017, 50(11):1941-1959.
doi: 10.3864/j.issn.0578-1752.2017.11.001 |
| [2] | Tilman D, Balzer C, Hill J, et al. Global food demand and the sustainable intensification of agriculture. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108 (50):20260-20264. |
| [3] | 闫亚飞. 河套灌区不同饲草生产性能与品质研究. 北京: 中国农业科学院, 2016. |
| [4] |
Iwata H. Genomic selection in plant breeding: from theory to practice. Briefings in Functional Genomics, 2010, 9(2):166-177.
doi: 10.1093/bfgp/elq001 |
| [5] |
Heffner E L, Lorenz A J, Jannink J L, et al. Plant breeding with genomic selection: gain per unit time and cost. Crop Science, 2010, 50(5):1681-1690.
doi: 10.2135/cropsci2009.11.0662 |
| [6] |
Crossa J, Pérez P, Hickey J M, et al. Genomic prediction in CIMMYT maize and wheat breeding program. Heredity, 2013, 112 (1):48.
doi: 10.1038/hdy.2013.16 |
| [7] |
Li D, Wang P, Gu R, et al. Genetic relatedness and the ratio of subpopulation-common alleles are related in genomic prediction across structured subpopulations in maize. Plant Breeding, 2019, 138(6):802-809.
doi: 10.1111/pbr.v138.6 |
| [8] | Xu S, Zhu D, Zhang Q. Predicting hybrid performance in rice using genomic best linear unbiased prediction. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(34):12456-12461. |
| [9] |
Cui Y, Li R, Li G, et al. Hybrid breeding of rice via genomic selection. Plant Biotechnology Journal, 2020, 18(1):57-67.
doi: 10.1111/pbi.13170 pmid: 31124256 |
| [10] |
Huang X, Yang S, Gong J, et al. Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nature Communications, 2015, 6(7052):6258.
doi: 10.1038/ncomms7258 |
| [11] | Nickolai A, Tai S, Wang W, et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Research, 2015 (D1):1023-1027. |
| [12] | Hadasch S, Simko I, Hayes R J, et al. Comparing the predictive abilities of phenotypic and marker-assisted selection methods in a biparental lettuce population. Plant Genome, 2016, 9(1):1-11. |
| [13] |
Yang J, Mezmouk S, Baumgarten A, et al. Incomplete dominance of deleterious alleles contributes substantially to trait variation and heterosis in maize. PLoS Genetics, 2016, 13(9):e1007019.
doi: 10.1371/journal.pgen.1007019 |
| [14] | Bates D, Mächler M, Bolker B, et al. Fitting linear mixed-effects models using lme4. Statistics & Computing, 2014, 1406(1):133-199. |
| [15] | Hallauer A R, Carena M J, Miranda Filho J B. Quantitative genetics in maize breeding. New York: Springer Science & Business Media, 2010. |
| [16] |
Murray M G, Thompson C L, Wendel J F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research, 1980, 8 (19):4321-4325.
doi: 10.1093/nar/8.19.4321 pmid: 7433111 |
| [17] |
Pérez P, Campos G D L. Genome-wide regression and prediction with the BGLR statistical package. Genetics, 2014, 198(2):483-495.
doi: 10.1534/genetics.114.164442 pmid: 25009151 |
| [18] |
Liu X, Wang H, Wang H, et al. Factors affecting genomic selection revealed by empirical evidence in maize. The Crop Journal, 2018, 6(4):341-352.
doi: 10.1016/j.cj.2018.03.005 |
| [19] |
Dias K O D G, Gezan S A, Guimarães C T, et al. Improving accuracies of genomic predictions for drought tolerance in maize by joint modeling of additive and dominance effects in multi- environment trials. Heredity, 2018, 121(1):24-37.
doi: 10.1038/s41437-018-0053-6 |
| [20] |
Massman J M, Gordillo A, Lorenzana R E, et al. Genomewide predictions from maize single-cross data. Theoretical and Applied Genetics, 2013, 126(1):13-22.
doi: 10.1007/s00122-012-1955-y pmid: 22886355 |
| [21] |
Hayes B J, Bowman P J, Chamberlain A C, et al. Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genetics Selection Evolution, 2009, 41(1):1-9.
doi: 10.1186/1297-9686-41-1 |
| [22] | 刘策, 孟焕文, 程智慧. 植物全基因组选择育种技术原理与研究进展. 分子植物育种, 2020, 18(16):5335-5342. |
| [23] | 陈雨, 姜淑琴, 孙炳蕊, 等. 基因组选择及其在作物育种中的应用. 广东农业科学, 2017, 44(9):1-7. |
| [1] | Cao Qingjun, Li Gang, Yang Hao, Lou Yuyong, Yang Fentuan, Kong Fanli, Li Xinbei, Zhao Xinkai, Jiang Xiaoli. The Effects of Different Tillage Practices on Seedbed Quality and Its Relationships with Seedling Population Construction and Grain Yield of Spring Maize [J]. Crops, 2023, 39(5): 249-254. |
| [2] | Yu Le, Li Lin, Huang Hongjuan, Huang Zhaofeng, Zhu Wenda, Wei Shouhui. Weed Species Composition and Community Characterization in Maize Fields in Hubei Province [J]. Crops, 2023, 39(5): 272-279. |
| [3] | Qu Haitao, Li Zhongnan, Wang Yueren, Ma Yiwen, Xiang Yang, Wu Shenghui, Tan Zhuo, Wang Chun, Wei Qiang, Luo Yao, Li Guangfa. Study on Genetic and Breeding Effects of 100-Grain Weight in Maize [J]. Crops, 2023, 39(5): 66-70. |
| [4] | Yang Mi, Wang Meijuan, Xu Haitao. Study on the Dynamic Development Difference of Husk of Maize Inbred Lines in Different Ecological Regions [J]. Crops, 2023, 39(5): 81-90. |
| [5] | Yuan Liuzheng, Wang Huiqiang, WangQiuling , Zhu Shidie, ZhaoYueqiang , Yuan Manman, Wang Huitao, Zhang Yundong, Liu Jiayou, Yuan Yongqiang. Analysis of Combining Ability and Genetic Effect of Maize Inbred Lines under Shading Condition [J]. Crops, 2023, 39(4): 104-109. |
| [6] | Zheng Fei, Chen Jing, Cui Yakun, Kong Lingjie, Meng Qingchang, Li Jie, Liu Ruixiang, Zhang Meijing, Zhao Wenming, Chen Yanping. Screening of High and Stable Yield Maize Varieties Suitable for Grain Mechanical Harvesting in Different Ecological Areas of the Huaibei Region [J]. Crops, 2023, 39(4): 110-117. |
| [7] | Wang Liping, Bai Lanfang, Wang Tianhao, Wang Xiaoxuan, Bai Yunhe, Wang Yufen. Effects of Different Nitrogen Levels on Nitrogen Accumulation and Transport in Silage Maize [J]. Crops, 2023, 39(4): 165-173. |
| [8] | Li Yuxin, Lu Min, Zhao Jiuran, Wang Ronghuan, Xu Tianjun, Lü Tianfang, Cai Wantao, Zhang Yong, Xue Honghe, Liu Yueʼe. The Production Status Investigation and Analysis of Summer Maize in Beijing-Tianjin-Tangshan Region [J]. Crops, 2023, 39(4): 174-181. |
| [9] | Liu Songtao, Tian Zaimin, Liu Zigang, Gao Zhijia, Zhang Jing, He Donggang, Huang Zhihong, Lan Xin. Transcriptomic Analysis to Reveal Lodging Resistance Genes and Metabolism Pathways in Maize (Zea mays L.) [J]. Crops, 2023, 39(4): 31-37. |
| [10] | Wen Shenghui, Yang Junwei, Wang Yang, Li Gongjian, Weng Jianfeng, Duan Canxing, Jia Xin, Wang Jianjun. Research Progress on Discovery of Resistance Genes and Molecular Breeding Utilization of Fungal Diseases in Maize [J]. Crops, 2023, 39(3): 1-11. |
| [11] | Chang Qing, Li Lijun, Qu Jiahui, Zhang Yanli, Han Dongyu, Zhao Xinyao. Yield Advantage and Nitrogen Use Efficiency of Forage Maize-Rape Intercropping Following Wheat in Tumed Plain [J]. Crops, 2023, 39(3): 167-174. |
| [12] | Guo Shulei, Wang Ying, Wei Liangming, Zhang Xin, Liu Yan, Wu Weihua, Lu Daowen, Lei Xiaobing, Wang Zhenhua, Lu Xiaomin. Analysis of Influencing Factors of Maize Yield under Different Ecological Conditions [J]. Crops, 2023, 39(3): 205-214. |
| [13] | Gao Mutian, Qiu Guanjie, Zhu Tongtong, Li Ruilian, Deng Min, Luo Hongbing, Huang Cheng. Dissecting the Genetic Basis of Flag Leaf in Maize-Teosinte Introgression Line Population [J]. Crops, 2023, 39(3): 51-57. |
| [14] | Li Zhongnan, Wang Yueren, Ma Yiwen, Xiang Yang, Wu Shenghui, Qu Haitao, Li Fulin, Zhang Shuqin, Li Guangfa. Genetic Analysis of Color Traits in Sheath, Silk, Anther and Cob of Isolated Population Based on Maize DH Lines [J]. Crops, 2023, 39(3): 75-79. |
| [15] | Zhang Panpan, Li Chuan, Zhang Meiwei, Zhao Xia, Huang Lu, Liu Jingbao, Qiao Jiangfang. Effects of Nitrification Inhibitor on the Nitrogen Concentration and Yield in Summer Maize Plants and Soil under Reduced Nitrogen Application [J]. Crops, 2023, 39(2): 145-150. |
|
||