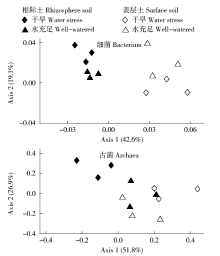
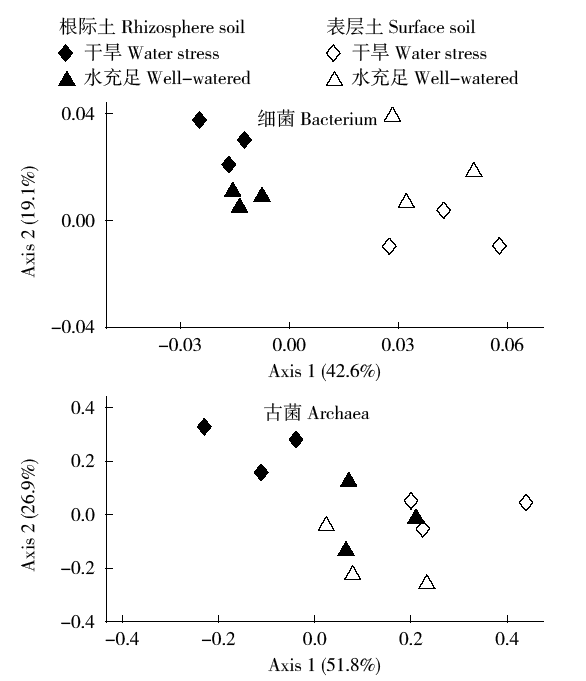
Crops ›› 2017, Vol. 33 ›› Issue (1): 127-134.doi: 10.16035/j.issn.1001-7283.2017.01.023
Previous Articles Next Articles
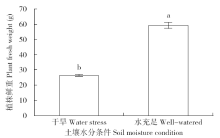
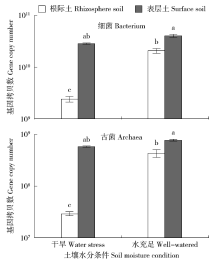
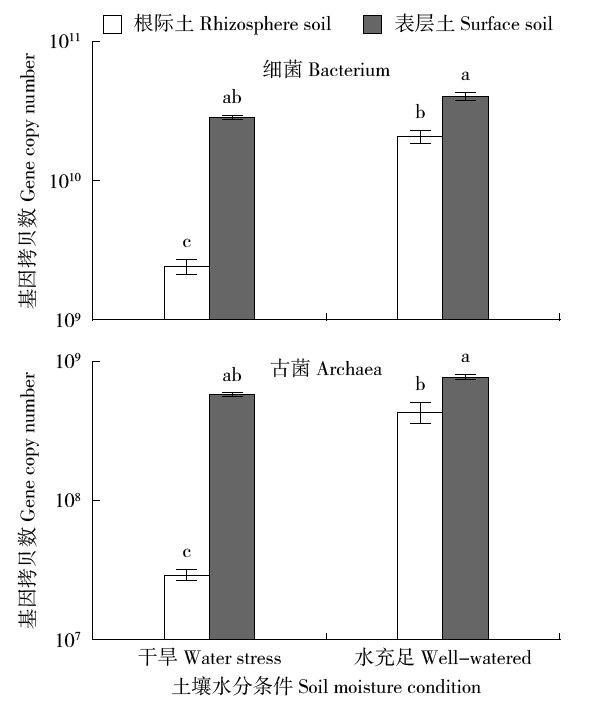
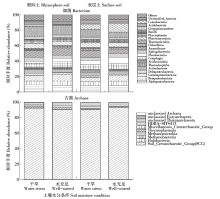
Effects of Water Conditions on Microbial Community in Maize Rhizosphere
Feng Shuai1,2,Liu Xiaoli1,2,Wu Xiaoli1,2,Jiang Shijie2,Zhou Zhengfu2,Zhang Wei2,Chen Ming2,Wang Jin1,2,Ke Xiubin2
- 1Life Science and Engineering College,Southwest University of Science and Technology,Mianyang 621010,Sichuan
2Biotechnology Research Institute,Chinese Academy of Agriculture Science,Beijing 100081,China
| [1] |
Bastida F, Moreno J L, Nicolás C , et al. Soil metaproteomics:a review of an emerging environmental science.Significance,methodology and perspectives. European Journal of Soil Science, 2009,60(6):845-859.
doi: 10.1111/j.1365-2389.2009.01184.x |
| [2] |
Chaudary D R, Gautam R K, Yousuf B , et al. Nutrients,microbial community structure and functional gene abundance of rhizosphere and bulk soils of halophytes. Applied Soil Ecology, 2015,91:16-26.
doi: 10.1016/j.apsoil.2015.02.003 |
| [3] |
Singh B K, Millard P, Whiteley A S , et al. Unravelling rhizosphere-microbial interactions:opportunities and limitations. Trends in Microbiology, 2004,12(8):386-393.
doi: 10.1016/j.tim.2004.06.008 |
| [4] | East R . Microbiome:Soil science comes to life. Nature, 2013,501(7468):18-19. |
| [5] |
Berendsen R L, Pieterse C M, Bakker P A . The rhizosphere microbiome and plant health. Trends in Plant Science, 2012,17(8):478-486.
doi: 10.1016/j.tplants.2012.04.001 |
| [6] | Bååth E, Berg B, Lohm U , et al. Effects of experimental acidification and liming on soil organisms and decomposition in a Scots pine forest. Pedobiologia, 1980,20:85-100. |
| [7] | Bais H P, Weir T L, Perryl L G , et al. The role of root exudates in rhizosphere interations with plants and other organisms.Annual Review of Plant Biology.Palo Alto; Annual Reviews. 2006,57:233-266. |
| [8] |
Bardgett R, Mawdsley J, Edwards S , et al. Plant species and nitrogen effects on soil biological properties of temperate upland grasslands. Functional Ecology, 1999,13(5):650-660.
doi: 10.1046/j.1365-2435.1999.00362.x |
| [9] |
Berg G, Smalla K . Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiology Ecology, 2009,68(1):1-13.
doi: 10.1111/fem.2009.68.issue-1 |
| [10] |
Cavaglieri L, Orlando J, Etcheverry M . Rhizosphere microbial community structure at different maize plant growth stages and root locations. Microbiological Research, 2009,164(4):391-399.
doi: 10.1016/j.micres.2007.03.006 |
| [11] |
Castellanos T, Dohrmann A B, Imfeld G , et al. Search of environmental descriptors to explain the variability of the bacterial diversity from maize rhizospheres across a regional scale. European Journal of Soil Biology, 2009,45(5):383-393.
doi: 10.1016/j.ejsobi.2009.07.006 |
| [12] | Correa-galeote D, Bedmar E J, Fernández-González A J , et al. Bacterial communities in the rhizosphere of amilaceous maize (Zea mays L.) as assessed by pyrosequencing. Front Plant Science, 2016,7:1016. |
| [13] |
覃潇敏, 郑毅, 汤利 , 等. 玉米与马铃薯间作对根际微生物群落结构和多样性的影响. 作物学报, 2015,41(6):919-928.
doi: 10.3724/SP.J.1006.2015.00919 |
| [14] | 叶慧香, 崔跃原, 宋新元 , 等. 转cry1Ie基因抗虫玉米对土壤中细菌群落结构的影响. 生物安全学报, 2015,24(1):64-71. |
| [15] |
van Dijk E L, Auger H, Jaszczyszyn Y , et al. Ten years of next-generation sequencing technology. Trends in Genetics, 2014,30(9):418-426.
doi: 10.1016/j.tig.2014.07.001 |
| [16] |
Walker N J . Real-time and quantitative PCR:applications to mechanism-based toxicology. Journal of Biochemical and Molecular Toxicology, 2001,15(3):121-127.
doi: 10.1002/(ISSN)1099-0461 |
| [17] |
Collavino M M, Tripp H J, Frank I E , et al. NifH pyrosequencing reveals the potential for location-specific soil chemistry to influence N2-fixing community dynamics. Environmental Microbiology, 2014,16(10):3211-3223.
doi: 10.1111/emi.2014.16.issue-10 |
| [18] |
Chelius M, Triplett E . The Diversity of archaea and bacteria in association with the roots of Zea mays L. Microbial Ecology, 2001,41(3):252-263.
doi: 10.1007/s002480000087 |
| [19] |
Peiffer J A, Spor A, Koren O , et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proceedings of the Nationa Academy of Sciences of the United States of America, 2013,110(16):6548-6553.
doi: 10.1073/pnas.1302837110 |
| [20] |
Cao P, Zhang L M, Shen J P , et al. Distribution and diversity of archaea communities in selected Chinese soils. FEMS Microbiology Ecology, 2012,80(1):146-158.
doi: 10.1111/j.1574-6941.2011.01280.x |
| [21] | Atlas R M . Use of microbial diversity measurements to assess environmental stress.Current Perspectives in Microbial Ecology American Society for Microbiology,Washington,DC, 1984: 540-545. |
| [22] | Schimel J . Ecosystem consequences of microbial diversity and community structure.Arctic and Alpine Biodiversity:Patterns, Causes and Ecosystem Consequences, 1995: 239-254. |
| [23] |
Uhlířová E, Elhottova E, Tříska J , et al. Physiology and microbial community structure in soil at extreme water content. Folia Microbiologica, 2005,50(2):161-166.
doi: 10.1007/BF02931466 |
| [24] |
Valentine D L . Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nature Reviews Microbiology, 2007,5(4):316-323.
doi: 10.1038/nrmicro1619 |
| [25] | Grinnell J . The niche-relationships of the California Thrasher. Auk, 1916,34(4):427-433. |
| [26] | 杨万勤, 宋光煜 . 土壤生态位及其应用. 世界科技研究与发展, 2001,23(2):21-23. |
| [27] |
Grayston S J, Wang S, Campbell C D , et al. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biology and Biochemistry, 1998,30(3):369-378.
doi: 10.1016/S0038-0717(97)00124-7 |
| [28] |
Li X, Rui J, Mao Y , et al. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biology and Biochemistry, 2014,68:392-401.
doi: 10.1016/j.soilbio.2013.10.017 |
| [29] |
Garbeva P, Vanveen J A, Vanelsas J D . Microbial diversity in soil:Selection of microbial populations by plant and soil type and implications for disease suppressiveness. Phytopathology, 2004,42:243-270.
doi: 10.1146/annurev.phyto.42.012604.135455 |
| [30] | Brimecombe M J, Deleij F A, Lynch J M . The effect of root exudates on rhizosphere microbial populations.The Rhizosphere,Biochemistry and Organic Substances at the Soil-plant Interface,Marcel Dekker,Inc, 2000: 95-140. |
| [31] |
Ke X, Angel R, Lu Y , et al. Niche differentiation of ammonia oxidizers and nitrite oxidizers in rice paddy soil. Environmental Microbiology, 2013,15(8):2275-2292.
doi: 10.1111/emi.2013.15.issue-8 |
| [32] |
Ke X, Lu Y, Conrad R . Different behaviour of methanogenic archaea and Thaumarchaeota in rice field microcosms. FEMS Microbiology Ecology, 2014,87(1):18-29.
doi: 10.1111/1574-6941.12188 |
| [33] |
Uroz S, Bu E M, Murat C , et al. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environmental Microbiology Reports, 2010,2(2):281-288.
doi: 10.1111/emi4.2010.2.issue-2 |
| [1] | Wei Zhang,Liangqun Wang,Yong Liu,Yanfang Hao,Wei Yang,Hongyan Bai,Bo Wu. Optimization of the Factors Related to the Efficiency of Agrobacterium-Mediated Transformation of Sorghum [J]. Crops, 2018, 34(1): 56-61. |
| [2] | Jingang Liang,Zhengguang Zhang. Advance on Effects of Genetically ModifiedCrops on Soil Ecosystems [J]. Crops, 2017, 33(4): 1-6. |
|