Crops ›› 2022, Vol. 38 ›› Issue (4): 37-45.doi: 10.16035/j.issn.1001-7283.2022.04.006
Previous Articles Next Articles
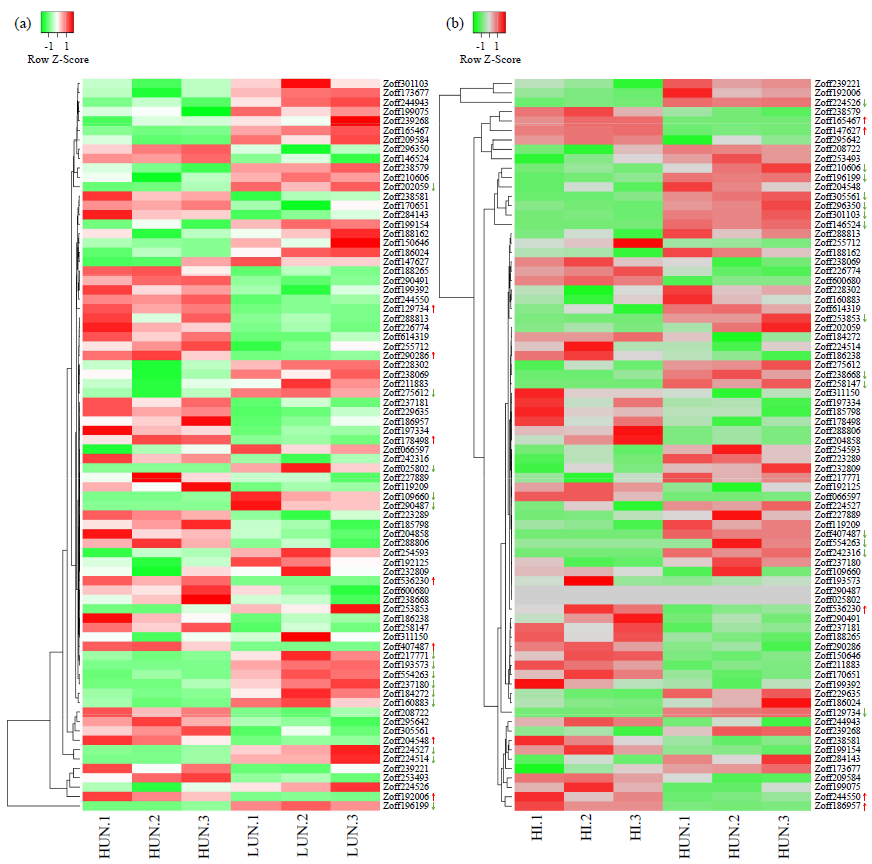
Identification and Expression Analysis of the ZoWRKY Family in Stress Responses Based on Transcriptome Data of Ginger (Zingiber officinale Roscoe)
Jiang Yusong1,2( ), Li Honglei2, Li Zhexin2, Xu Xiaoyu1, Li Longyun3, Huang Mengjun1,2,3(
), Li Honglei2, Li Zhexin2, Xu Xiaoyu1, Li Longyun3, Huang Mengjun1,2,3( )
)
- 1College of Resources and Environment, Southwest University, Chongqing 400715, China
2Institute for Special Plants, Chongqing University of Arts and Sciences, Chongqing 402160, China
3Institute of Chinese Materia Medica, Chongqing Academy of Chinese Materia Medica, Chongqing 400065, China
| [1] |
Allan A C, Hellens R P, Laing W A. MYB transcription factors that colour our fruit. Trends in Plant Science, 2008, 13(3):99-102.
doi: 10.1016/j.tplants.2007.11.012 |
| [2] |
Liu J, Osbourn A, Ma P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Molecular Plant, 2015, 8(5):689-708.
doi: 10.1016/j.molp.2015.03.012 |
| [3] |
Welling A, Palva E T. Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiology, 2008, 147(3):1199-1211.
doi: 10.1104/pp.108.117812 |
| [4] |
Zhao C, Zhang Z, Xie S, et al. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiology, 2016, 171(4):2744-2759.
doi: 10.1104/pp.16.00533 |
| [5] |
Olsen A N, Ernst H A, Leggio L L, et al. NAC transcription factors:structurally distinct,functionally diverse. Trends in Plant Science, 2005, 10(2):79-87.
doi: 10.1016/j.tplants.2004.12.010 |
| [6] | Zhang Z, Dong J, Ji C, et al. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proceedings of National Academy of Science of USA, 2019, 116(23):11223-11228. |
| [7] |
黄幸, 丁峰, 彭宏祥, 等. 植物WRKY转录因子家族研究进展. 生物技术通报, 2019, 35(12):129-143.
doi: 10.13560/j.cnki.biotech.bull.1985.2019-0626 |
| [8] |
Wu K L, Guo Z J, Wang H H, et al. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Research, 2005, 12(1):9-26.
doi: 10.1093/dnares/12.1.9 |
| [9] |
Rushton P J, Somssich I E, Ringler P, et al. WRKY transcription factors. Trends in Plant Science, 2010, 15(5):247-258.
doi: 10.1016/j.tplants.2010.02.006 pmid: 20304701 |
| [10] |
Eulgem T, Rushton P J, Robatzek S, et al. The WRKY superfamily of plant transcription factors. Trends in Plant Science, 2000, 5(5):199-206.
pmid: 10785665 |
| [11] | Chen L, Zhang L, Li D, et al. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proceedings of National Academy of Science of USA, 2013, 110(21):1963-1971. |
| [12] |
Jiang J, Ma S, Ye N, et al. WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology, 2017, 59(2):86-101.
doi: 10.1111/jipb.12513 |
| [13] |
Dang F F, Wang Y N, Yu L, et al. CaWRKY40,a WRKY protein of pepper,plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant,Cell and Environment, 2013, 36(4):757-774.
doi: 10.1111/pce.12011 |
| [14] |
Merz P R, Moser T, Holl J, et al. The transcription factor VvWRKY 33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmopara viticola. Physiologia Plantarum, 2015, 153(3):365-380.
doi: 10.1111/ppl.12251 |
| [15] | Cormack R S, Eulgem T, Rushton P J, et al. Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochimica Biophysica Acta, 2002, 1576(1):92-100. |
| [16] |
Bakshi M, Oelmuller R. WRKY transcription factors:Jack of many trades in plants. Plant Signaling and Behavior, 2014, 9(2):e27700.
doi: 10.4161/psb.27700 |
| [17] | 徐惠娟, 郑蕊, 陈任, 等. 枸杞WRKY3基因克隆及组织表达分析. 西北植物学报, 2016, 36(9):1721-1727. |
| [18] |
Cai H, Yang S, Yan Y, et al. CaWRKY6 transcriptionally activates CaWRKY40,regulates Ralstonia solanacearum resistance,and confers high-temperature and high-humidity tolerance in pepper. Journal of Experimental Botany, 2015, 66(11):3163-3174.
doi: 10.1093/jxb/erv125 |
| [19] | 李可, 熊茜, 肖晓蓉, 等. 木薯25个WRKY家族转录因子在生物胁迫下的表达分析. 热带生物学报, 2017, 8(1):14-21. |
| [20] |
Ülker B, Somssich I E. WRKY transcription factors:from DNA binding towards biological function. Current Opinion in Plant Biology, 2004, 7(5):491-498.
doi: 10.1016/j.pbi.2004.07.012 |
| [21] |
Ross C A, Liu Y, Shen Q J. The WRKY gene family in rice (Oryza sativa). Journal of Integrative Plant Biology, 2007, 49(6):827-842.
doi: 10.1111/j.1744-7909.2007.00504.x |
| [22] |
Huang S, Gao Y, Liu J, et al. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Molecular Genetics and Genomics, 2012, 287(6):495-513.
doi: 10.1007/s00438-012-0696-6 |
| [23] |
Dou L, Zhang X, Pang C, et al. Genome-wide analysis of the WRKY gene family in cotton. Molecular Genetics and Genomics, 2014, 289(6):1103-1121.
doi: 10.1007/s00438-014-0872-y |
| [24] |
Semwal R B, Semwal D K, Combrinck S, et al. Gingerols and shogaols:important nutraceutical principles from ginger. Phytochemistry, 2015, 117:554-568.
doi: 10.1016/j.phytochem.2015.07.012 |
| [25] |
Sun C, Palmqvist S, Olsson H, et al. A novel WRKY transcription factor,SUSIBA2,participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. The Plant Cell, 2003, 15(9):2076-2092.
doi: 10.1105/tpc.014597 |
| [26] | 谷彦冰, 冀志蕊, 迟福梅, 等. 苹果WRKY基因家族生物信息学及表达分析. 中国农业科学, 2015, 48(16):3221-3238. |
| [27] |
Eulgem T, Somssich I E. Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology, 2007, 10(4):366-371.
doi: 10.1016/j.pbi.2007.04.020 |
| [28] | 向小华, 吴新儒, 晁江涛, 等. 普通烟草WRKY基因家族的鉴定及表达分析. 遗传, 2016, 38(9):840-856. |
| [29] |
Ali M A, Azeem F, Nawaz M A, et al. Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. Journal of Plant Physiology, 2018, 226:12-21.
doi: 10.1016/j.jplph.2018.04.007 |
| [30] |
Li J, Brader G, Kariola T, et al. WRKY70 modulates the selection of signaling pathways in plant defense. The Plant Journal, 2006, 46(3):477-491.
doi: 10.1111/j.1365-313X.2006.02712.x |
| [31] |
Chen X, Liu J, Lin G, et al. Overexpression of AtWRKY28 and AtWRKY 75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Reports, 2013, 32(10):1589-1599.
doi: 10.1007/s00299-013-1469-3 |
| [32] |
Scarpeci T E, Zanor M I, Mueller-Roeber B, et al. Overexpression of AtWRKY 30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Molecular Biology, 2013, 83(3):265-277.
doi: 10.1007/s11103-013-0090-8 pmid: 23794142 |
| [33] |
Xu X, Chen C, Fan B, et al. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40,and WRKY 60 transcription factors. The Plant Cell, 2006, 18(5):1310-1326.
doi: 10.1105/tpc.105.037523 |
| [1] | Wang Tong, Zhao Xiaodong, Zhen Pingping, Chen Jing, Chen Mingna, Chen Na, Pan Lijuan, Wang Mian, Xu Jing, Yu Shanlin, Chi Xiaoyuan, Zhang Jiancheng. Genome-Wide Identification and Characteristic Analyzation of the TCP Transcription Factors Family in Peanut [J]. Crops, 2021, 37(2): 35-44. |
| [2] | Li Guolong, Wu Haixia, Sun Yaqing. Construction of RNAi Expression Vector of BvWRKY23 Gene in Sugar Beet [J]. Crops, 2020, 36(5): 41-47. |
| [3] | Xu Yuanyuan, Zhao Peng, Hong Quanchun, Zhu Xiaoqin, Pei Dongli. Isolation and Expression Analysis of Transcription Factor Gene TaMYB70 in Wheat [J]. Crops, 2020, 36(4): 84-90. |
| [4] | Duan Junzhi, Qi Xueli, Feng Lili, Zhang Huifang, Sun Yan, Yan Zhaoling, Chen Haiyan, Qi Hongzhi, Fan Wenjie, Yang Cuiping, Liu Yuxia, Ren Yinling, Zhang Jiayuan, Li Ying, Zhuo Wenfei. Progress on Application of Drought Tolerance Genes in Wheat Drought Tolerance Genetic Engineering [J]. Crops, 2020, 36(3): 7-15. |
| [5] | Yang Junkai,Shen Yang,Cai Xiaoxi,Wu Shengyang,Li Jianwei,Sun Mingzhe,Jia Bowei,Sun Xiaoli. Genome-Wide Identification and Expression Patterns Analysis of the PHD Family Protein in Glycine max [J]. Crops, 2019, 35(3): 55-65. |
| [6] | Wu Hao,Li Yanmin,Xie Chuanxiao. Research Advances on Physiological Basis and Gene Discovery for Thermal Tolerance in Crops [J]. Crops, 2018, 34(5): 1-9. |
| [7] | Ying Zhang,Pengyu Liu,Xue Bai,Yang Yang,Yueying Li. Expression and Bioinformatics Analysis of CsWRKY23 Gene in Cucumber [J]. Crops, 2017, 33(5): 38-42. |
| [8] | Xue Zhang,Yuejia Yin,Bei Fan,Huijie Li,Xiaoyu Fei,Xiyan Cui. Advances on the Structural Characteristics and Function of Dof Transcription Factors in Plant [J]. Crops, 2016, 32(2): 14-20. |
|
||