Crops ›› 2019, Vol. 35 ›› Issue (3): 55-65.doi: 10.16035/j.issn.1001-7283.2019.03.010
Previous Articles Next Articles
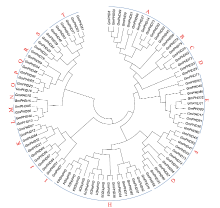
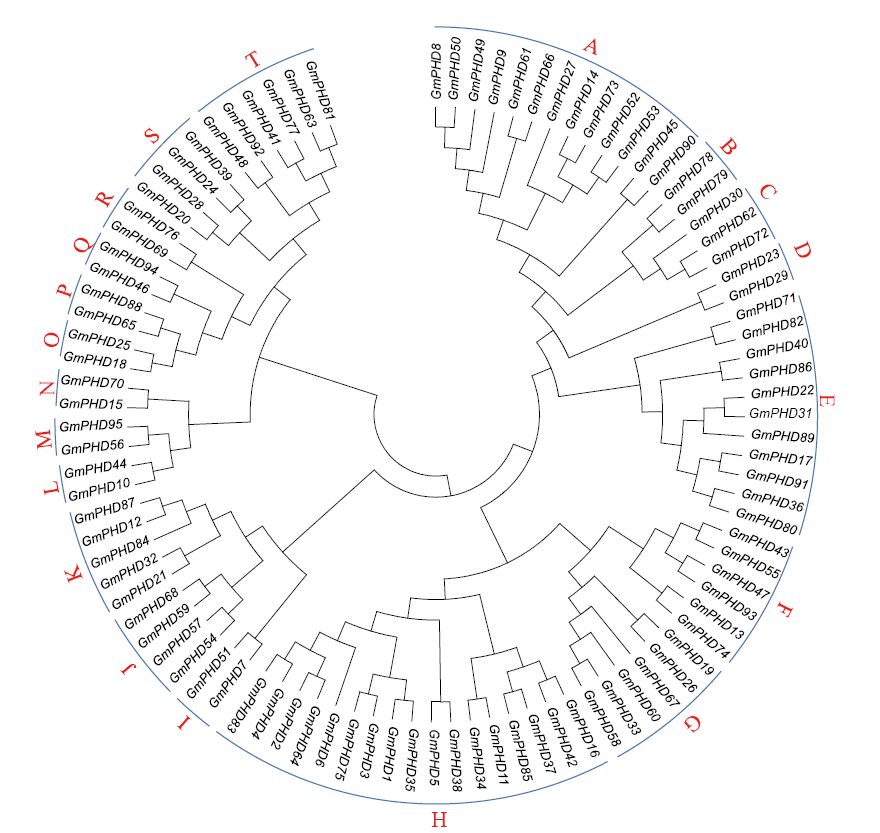
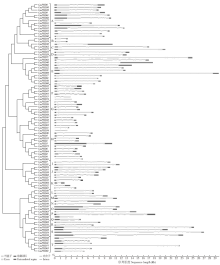
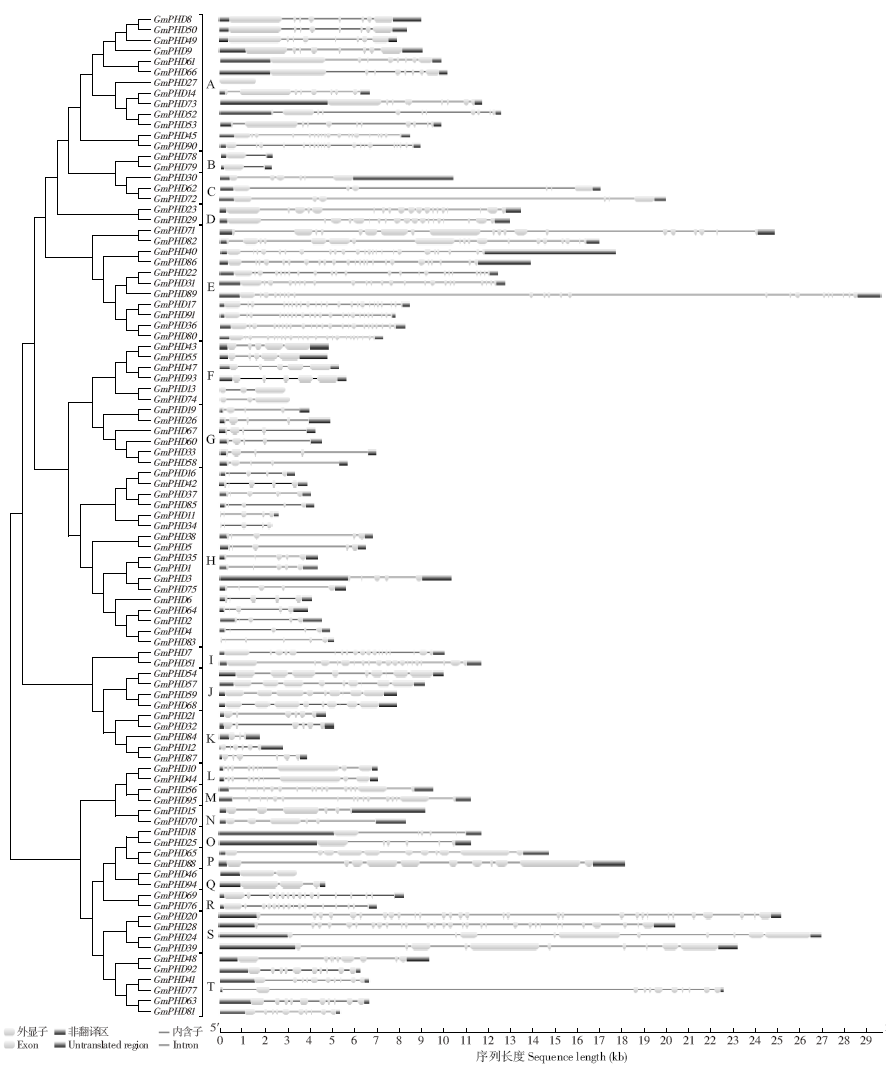
Genome-Wide Identification and Expression Patterns Analysis of the PHD Family Protein in Glycine max
Yang Junkai,Shen Yang,Cai Xiaoxi,Wu Shengyang,Li Jianwei,Sun Mingzhe,Jia Bowei,Sun Xiaoli
- College of Agronomy, Heilongjiang Bayi Agricultural University/Crop Stress Molecular Biology Laboratory, Daqing 163319, Heilongjiang, China
| [1] |
韩丹, 王丕武, 曲静 , 等. 大豆C2H2型锌指蛋白基因SCTF-1转化及功能分析. 中国油料作物学报, 2016,38(3):307-312.
doi: 10.7505/j.issn.1007-9084.2016.03.006 |
| [2] |
Agarwal P, Arora R, Ray S , et al. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Molecular Biology, 2007,65(4):467-485.
doi: 10.1007/s11103-007-9199-y |
| [3] |
Hall T M . Multiple modes of RNA recognition by zinc finger proteins. Current Opinion in Structure Biology, 2005,15(3):367-373.
doi: 10.1016/j.sbi.2005.04.004 |
| [4] |
Arnaud D, Dejardin A, Leple J G , et al. Genome-wide analysis of LIM gene family in Populus trichocarpa,Arabidopsis thaliana,and Oryza sativa. DNA Research, 2007,14(3):103-116.
doi: 10.1093/dnares/dsm013 |
| [5] |
Takatsuji H . Zinc-finger transcription factors in plants. Cellular and Molecular Life Sciences, 1998,54:582-596.
doi: 10.1007/s000180050186 |
| [6] |
Aasland R, Gibson T J, Stewart A F . The PHD finger:Implications for chromatin-mediated transcriptional regulation. Trends in Biochemical Sciences, 1995,20:56-59.
doi: 10.1016/S0968-0004(00)88957-4 |
| [7] |
Borden K L B, Freemont S P . The ring finger domain:A recent example of a sequence-structure family. Current Opinion in Structural Biology, 1996,6:395-401.
doi: 10.1016/S0959-440X(96)80060-1 |
| [8] |
Kaadige M R, Ayer D E . The polybasic region that follows the plant homeodomain zinc finger 1 of Pf1 is necessary and sufficient for specific phosphoinositide binding. Journal of Biological Chemistry, 2006,281(39):28831-28836.
doi: 10.1074/jbc.M605624200 |
| [9] | 冯英, 刘庆坡, 薛庆中 . 水稻与拟南芥PHD-finger蛋白的系统分析. 遗传学报, 2004,31(11):1284-1293. |
| [10] |
Wang Q, Liu J, Wang Y , et al. Systematic analysis of the maize PHD-finger gene family reveals a subfamily involved in abiotic stress response. International Journal of Molecular Sciences, 2015,16(10):23517-23544.
doi: 10.3390/ijms161023517 |
| [11] |
Wu S, Wu M, Dong Q , et al. Genome-wide identification,classification and expression analysis of the PHD-finger protein family in Populus trichocarpa. Gene, 2016,575(1):75-89.
doi: 10.1016/j.gene.2015.08.042 |
| [12] |
Sun M Z, Jia B W, Yang J K , et al. Genome-wide identification of the PHD-finger family genes and their responses to environmental stresses in Oryza sativa L. International Journal of Molecular Sciences, 2017,18(9):2005.
doi: 10.3390/ijms18092005 |
| [13] | Chen X J, Sikandar H, Cheng Z H . Genome-wide identification and analysis of genes encoding PHD-finger protein in tomato. Pakistan Journal of Botany, 2016,48(1):155-165. |
| [14] | Wu X J, Li M Y, Que F , et al. Genome-wide analysis of PHD family transcription factors in carrot (Daucus carota L.) reveals evolution and response to abiotic stress. Acta Physiologiae Plantarum, 2016,38(3):2049-2061. |
| [15] | 张帆, 杨仲南, 张森 . 拟南芥PHD-finger-蛋白家族的全基因组分析. 云南植物研究, 2009,31(3):227-238. |
| [16] |
Ye Y, Gong Z, Lu X , et al. Germostatin resistance locus 1 encodes a PHD finger protein involved in auxin-mediated seed dormancy and germination. Plant Journal, 2016,85(1):3-15.
doi: 10.1111/tpj.13086 |
| [17] |
Kim D H, Sung S . The binding specificity of the PHD-finger domain of VIN3 moderates vernalization response. Plant Physiology, 2017,173(2):1258-1268.
doi: 10.1104/pp.16.01320 |
| [18] |
Fernandez Gomez J, Wilson Z A . A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotechnology Journal, 2014,12(6):765-777.
doi: 10.1111/pbi.2014.12.issue-6 |
| [19] |
Lopez-Gonzalez L, Mouriz A, Narro-Diego L , et al. Chromatin-dependent repression of the Arabidopsis floral integrator genes involves plant specific PHD-containing proteins. The Plant Cell, 2014,26(10):3922-3938.
doi: 10.1105/tpc.114.130781 |
| [20] |
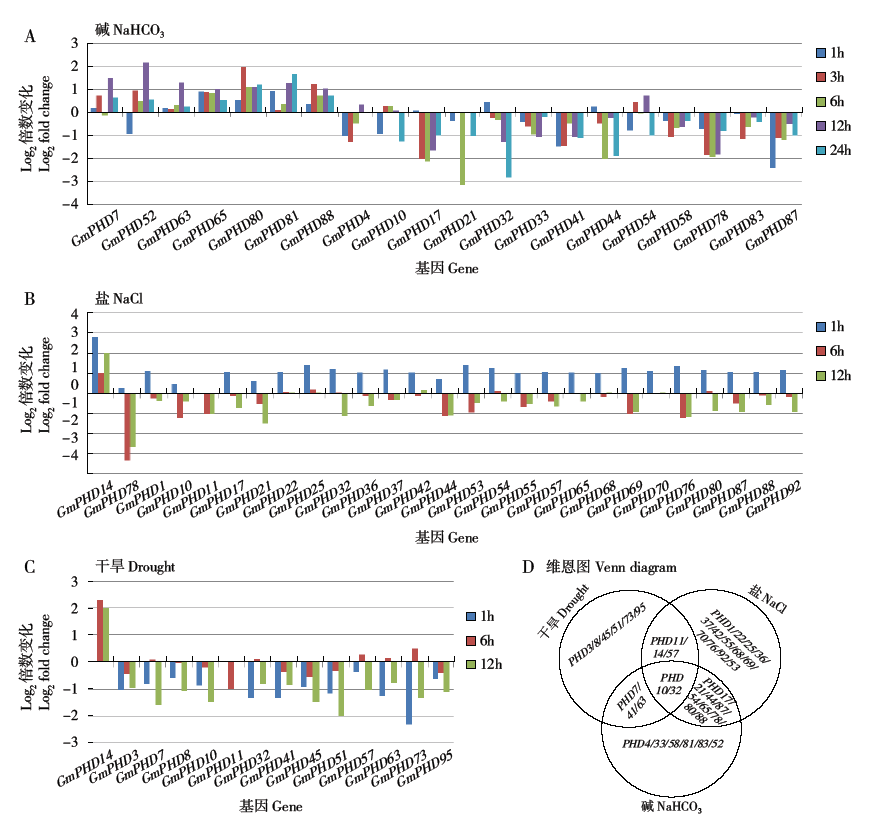
Wei W, Huang J, Hao Y J , et al. Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS ONE, 2009,4(9):e7209.
doi: 10.1371/journal.pone.0007209 |
| [21] |
Wu T, Pi E X, Tsai S N , et al. GmPHD5 acts as an important regulator for crosstalk between histone H3K4 di-methylation and H3K14 acetylation in response to salinity stress in soybean. BMC Plant Biology, 2011,11:178.
doi: 10.1186/1471-2229-11-178 |
| [22] |
Zhang H, Meltzer P, Davis S . RCircos:an R package for Circos 2D track plots. BMC Bioinformatics, 2013,14:244.
doi: 10.1186/1471-2105-14-244 |
| [23] | 李晓旭, 刘成, 李伟 , 等. 番茄WOX转录因子家族的鉴定及进化、表达分析. 遗传, 2016,38(5):444-460. |
| [24] |
Schmutz J, Cannon S B, Schlueter J , et al. Genome sequence of the palaeopolyploid soybean. Nature, 2010,463:178-183.
doi: 10.1038/nature08670 |
| [25] |
Winicov I . Alfin1 transcription factor overexpression enhances plant root growth under normal and saline conditions and improves salt tolerance in alfalfa. Planta, 2000,210(3):416-422.
doi: 10.1007/PL00008150 |
| [26] |
Winicov I I, Bastola D R . Transgenic overexpression of the transcription factor alfin1 enhances expression of the endogenous MsPRP2 gene in alfalfa and improves salinity tolerance of the plants. Plant Physiology, 1999,120(2):473-480.
doi: 10.1104/pp.120.2.473 |
| [27] |
Robert-Seilaniantz A, Grant M, Jones J D . Hormone crosstalk in plant disease and defense:more than just jasmonate-salicylate antagonism. Annual Review of Phytopathology, 2011,49:317-343.
doi: 10.1146/annurev-phyto-073009-114447 |
| [28] |
Callebaut I, Courvalin J C, Mornon J P . The BAH (bromo-adjacent homology) domain:a link between DNA methylation,replication and transcriptional regulation. FEBS Letters, 1999,446(1):189-193.
doi: 10.1016/S0014-5793(99)00132-5 |
| [29] |
Oliver A W, Jones S A, Roe S M , et al. Crystal structure of the proximal BAH domain of the polybromo protein. Biochemical Journal, 2005,389(3):657-664.
doi: 10.1042/BJ20050310 |
| [30] |
Baumbusch L O, Thorstensen T, Krauss V , et al. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Research, 2001,29(21):4319-4333.
doi: 10.1093/nar/29.21.4319 |
| [31] |
Cui X, De Vivo I, Slany R , et al. Association of SET domain and myotubularin-related proteins modulates growth control. Nature Genetics, 1998,18(4):331-337.
doi: 10.1038/ng0498-331 |
| [32] |
Stec I, Nagl S B, van Ommen G J , et al. The PWWP domain:a potential protein-protein interaction domain in nuclear proteins influencing differentiation. FEBS Letters, 2000,473(1):1-5.
doi: 10.1016/S0014-5793(00)01449-6 |
| [33] |
Qin S, Min J R . Structure and function of the nucleosome-binding PWWP domain. Trends in Biochemical Sciences, 2014,39(11):536-547.
doi: 10.1016/j.tibs.2014.09.001 |
| [34] |
Molitor A M, Bu Z Y, Yu Y , et al. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genetics, 2014,10(1):e1004091.
doi: 10.1371/journal.pgen.1004091 |
| [1] | Xixi Dai,Heming Zhan,Xinghong Cui,Yinyue Zhao,Dandan Shan,Liang Zhang,Tiejun Wang. A Mathematical Model of Density Coupling and Its Optimization in Maize-Soybean Intercropping [J]. Crops, 2019, 35(2): 128-135. |
| [2] | Chunyu Lin,Xiaoyu Liang,Huiyan Zhao,Yang Wang. Analysis of Genetic Diversity and Population Structure of Main Soybean Varieties in Heilongjiang Province [J]. Crops, 2019, 35(2): 78-83. |
| [3] | Bo Liu,Ling Wei,Junhong Xiao,Haifeng Yang,Xueyan Duan,Aiping Chen,Ruilan Ren. Study on Improving the Hybrid Seed Setting Rate of Soybean [J]. Crops, 2019, 35(1): 81-84. |
| [4] | Yue Li,Haiyan Li,Jidong Yu,Jie Deng,Yuanfu Gong,Junshu Zhu. Allelopathy of Extracts from Hemp Straw on Soybean [J]. Crops, 2019, 35(1): 175-179. |
| [5] | Yun Zhao,Cailong Xu,Xu Yang,Suzhen Li,Jing Zhou,Jicun Li,Tianfu Han,Cunxiang Wu. Effects of Sowing Methods on Seedling Stand and Production Profit of Summer Soybean under Wheat-Soybean System [J]. Crops, 2018, 34(4): 114-120. |
| [6] | Mingjun Zhang,Zhongfeng Li,Lili Yu,Jun Wang,Lijuan Qiu. Identification and Screening of Protein Subunit Variation Germplasm from Both Mutants and Natural Population in Soybean [J]. Crops, 2018, 34(3): 44-50. |
| [7] | Tianle Ma,Jianxin Zhang. Effects of Different Multiple Cropping Methods on Dry Matter Accumulation, Distribution and Yield of Summer Soybean [J]. Crops, 2018, 34(1): 156-159. |
| [8] | Jiani Zhu,Huiping Dai,Shuhe Wei,Genliang Jia,Dejing Chen,Jinjin Pei,Qing Zhang,Long Qiang. Effects of Applying Zn on the Growth and Zn Accumulation in Soybean at Flowering Stage [J]. Crops, 2018, 34(1): 152-155. |
| [9] | Lina Li,Longguo Jin,Chuanxiao Xie,Changlin Liu. Determining Blind Samples of Transgenic Maize and Transgenic Soybean [J]. Crops, 2017, 33(6): 37-44. |
| [10] | Xuechao Zhou,Surong Ding,Yunshan Wei,Yanfang Zhou,Xue Wei,Rina Na,Feng Li. Evaluation of Adaptability of Different Vegetable Soybean Cultivars (Lines) in Chifeng Area [J]. Crops, 2017, 33(3): 44-48. |
| [11] | Guoyong Ren,Wei Li,Lifeng Zhang,Caijie Wang,Haiying Dai,Jinlong Wang,Ran Xu,Yanwei Zhang. The Resistance to Soybean Cyst Nematode Race 1 of Transgenic Soybean with hrf2 Encoding HarpinXooc [J]. Crops, 2017, 33(3): 49-53. |
| [12] | Xuli Zhang,Baolong Xing,Guimei Wang,Lili Yin. Effects of Planting Density on Agronomic Traits, Economic Traits and Yield of Soybean in North of Shanxi Province [J]. Crops, 2017, 33(3): 127-131. |
| [13] | Xiting Zhang,Liwei Cao,Shucai Lü,Guoxing Chen,Yongji Wang,Shuhan Yu,Zhenping Gong. Effects of Bulk Density on Nitrogen Absorption and Yield of Soybean on Black Soil [J]. Crops, 2017, 33(3): 132-137. |
| [14] | Li Yan,Qiang Yang,Yupeng Shao,Dandan Li,Zhikun Wang,Wenbin Li. Cloning and Sequence Analysis of GmWRI1a Gene Promoter in Soybean [J]. Crops, 2017, 33(2): 51-58. |
| [15] | Yixin Tian,Fengju Gao. The Response of Growth and Dry Matter Accumulation and Distribution of High Protein Soybean to Plant Density [J]. Crops, 2017, 33(2): 121-125. |
|