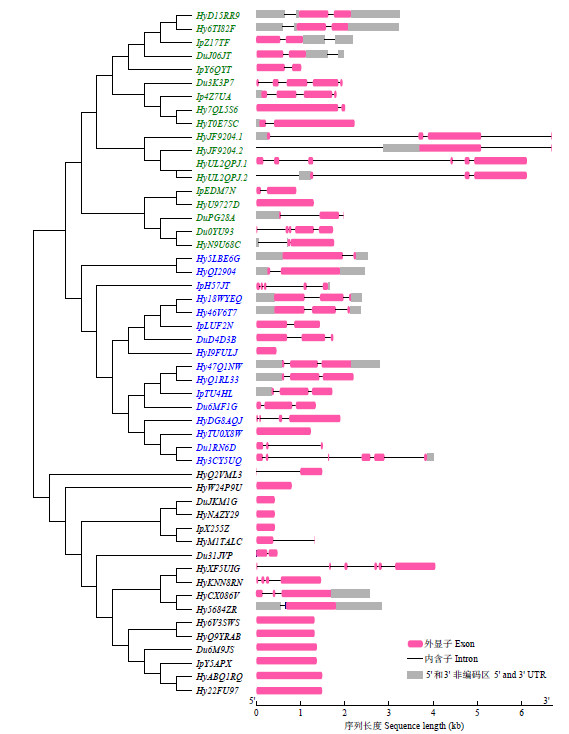
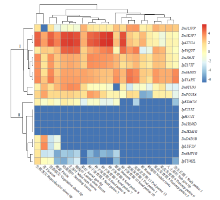
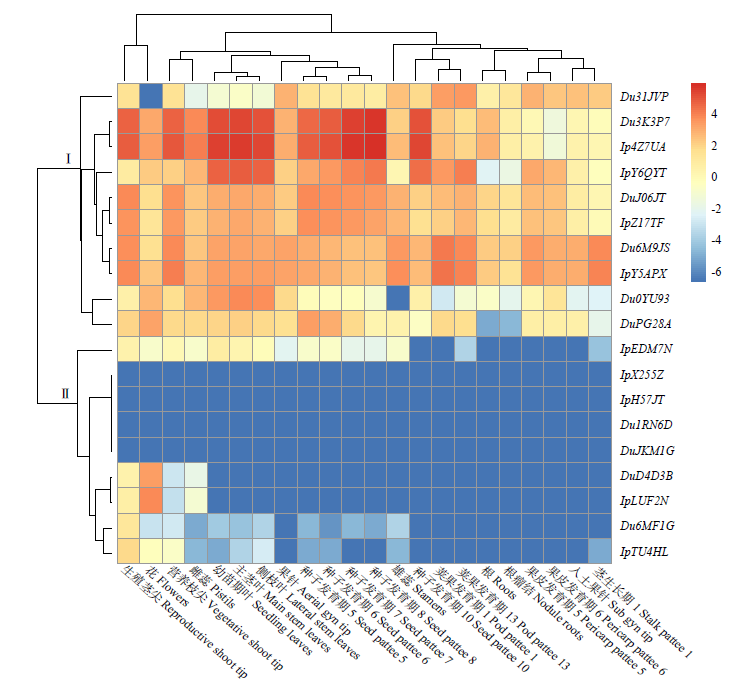
| [1] |
Cubas P, Lauter N, Doebley J, et al. The TCP domain:a motif found in proteins regulating plant growth and development. The Plant Journal, 1999,18(2):215-222.
|
| [2] |
Wang H F, Wang H W, Liu R, et al. Genome-wide identification of TCP family transcription factors in Medicago truncatula reveals significant roles of miR319-targeted TCPs in nodule development. Frontiers in Plant Science, 2018,9:774.
|
| [3] |
Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize, Nature, 1997,386(6624):485-488.
|
| [4] |
Luo D, Carpenter R, Vincent C, et al. Origin of floral asymmetry in Antirrhinum. Nature, 1996,383(6603):794-799.
|
| [5] |
Aggarwal P, Das Gupta M, Joseph A P. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell, 2010,22:1174-1189.
|
| [6] |
Dhaka N, Bhardwaj V, Sharma M K, et al. Evolving tale of TCPs:new paradigms and old lacunae. Frontiers in Plant Science, 2017,8:479.
|
| [7] |
Martín-Trillo M, Cubas P. TCP genes:a family snapshot ten years later. Trends in Plant Science, 2010,15(1):31-39.
|
| [8] |
Zhao J, Zhai Z, Li Y, et al. Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.). Frontiers in Plant Science, 2018,9:1282.
|
| [9] |
Nicolas M, Rodriguezbuey M L, Francozorrilla J M, et al. A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Current Biology, 2015,25(14):1799-1809.
|
| [10] |
Kieffer M, Master V, Waites R, et al. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. The Plant Journal, 2011,68:147-158.
|
| [11] |
Nag A, King S, Jack T. MiR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 2009,106(52):22534-22539.
|
| [12] |
Resentini F, Felipo-Benavent A, Colombo L, et al. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Molecular Plant, 2015,8(3):482-485.
|
| [13] |
Gonzalez-Grandio E, Pajoro A, Franco Zorrilla J M, et al. Abscisic acid signaling is controlled by a BRANCHED1/H D-ZIP I cascade in Arabidopsis axillary buds. Proceedings of the National Academy of Sciences of the United States of America, 2017,114(2):E245-E254.
|
| [14] |
Zhou M, Li D Y, Li Z G, et al. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiology, 2013,161:1375-1391.
|
| [15] |
Liu H, Wu M, Li F, et al. TCP Transcription factors in moso bamboo (Phyllostachys edulis):genome-wide identification and expression analysis. Frontiers in Plant Science, 2018,9:1263.
|
| [16] |
Koyama T, Furutani M, Tasaka M, et al. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. The Plant Cell, 2007,19(2):473-484.
|
| [17] |
Takeda T, Amano K, Ohto M A, et al. RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Molecular Biology, 2006,61:165-177.
|
| [18] |
Zheng K, Ni Z, Qu Y, et al. Genome-wide identification and expression analyses of TCP transcription factor genes in Gossypium barbadense. Scientific Reports, 2018,8(1):14526.
|
| [19] |
Aguilar-Martinez J A, Poza-Carrion C, Cubas P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell, 2007,19:458-472.
|
| [20] |
Martin-Trillo M, Grandio E G, Serra F, et al. Role of tomato BRANCHED1- like genes in the control of shoot branching. Plant Journal, 2011,67(4):701-714.
|
| [21] |
刘洋, 张慧, 辛大伟, 等. 大豆TCP转录因子家族结构域分析及功能预测. 大豆科学, 2012,31(5):707-713,717.
|
| [22] |
Parapunova V, Busscher M, Busscher-Lange J, et al. Identification,cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biology, 2014,14(1):157.
|
| [23] |
Francis A, Dhaka N, Bakshi M, et al. Comparative phylogenomic analysis provides insights into TCP gene functions in Sorghum. Scientific Reports, 2016,6:38488.
|
| [24] |
Chai W, Jiang P, Huang G, et al. Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiology and Molecular Biology of Plants, 2017,23:779-791.
|
| [25] |
Leng X, Wei H, Xu X, et al. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genomics, 2019,20(1):786-786.
|
| [26] |
Bertiolid J, C annon S B, F roenicke L, et al. The genome sequences of Arachis duranensis and Arachis ipaensis,the diploid ancestors of cultivated peanut. Nature Genetics, 2016,48(4):438.
|
| [27] |
Zhuang W, Chen H, Yang M, et al. The genome of cultivated peanut provides insight into legume karyotypes,polyploid evolution and crop domestication. Nature Genetics, 2019,51(5):865-876.
|
| [28] |
Xu R, Sun P, Jia F, et al. Genomewide analysis of TCP transcription factor gene family in Malus domestica. Journal of Genetics, 2014,93(3):733-746.
|
| [29] |
王亚鹏. 马铃薯StTCP家族基因鉴定及其对块茎休眠解除的响应. 兰州:甘肃农业大学, 2019.
|
| [30] |
赵一彤, 魏戏梦, 薛超玲, 等. 枣TCP家族基因鉴定及其表达分析. 河北农业大学学报, 2019,42(5):39-45.
|
| [31] |
Li W, Li D D, Han L H, et al. Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum). Scientific Reports, 2017,7(1):10118.
|
| [32] |
华方静, 刘风珍, 万勇善, 等. 花生及其野生种质溶血磷脂酸酰基转移酶基因(LPAAT)的克隆及序列分析. 分子植物育种, 2014,12(1):74-79.
|
| [33] |
Liu Y, Guan X, Liu S, et al. Genome-wide identification and analysis of TCP transcription factors involved in the formation of leafy head in Chinese cabbage. International Journal of Molecular Sciences, 2018,19(3):847.
|
| [34] |
Huo Y Z, Xiong W D, Su K L, et al. Genome-Wide Analysis of the TCP Gene Family in Switchgrass(Panicum virgatum L.). International Journal of Genomics, 2019(1):1-13.
|
| [35] |
Feng K, Hao J, Liu J, et al. Genome-wide identification,classification,and expression analysis of TCP transcription factors in carrot. Canadian Journal of Plant Science, 2019,99(4):525-535.
|
| [36] |
Hubbard L, Mcsteen P, Doebley J, et al. Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics, 2002,162(4):1927-1935.
|
| [37] |
Finlayson S A. Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant and Cell Physiology, 2007,48(5):667-677.
|
| [38] |
Luo D, Carpenter R, Copsey L, et al. Control of organ asymmetry in flowers of Antirrhinum. Cell, 1999,99(4):367-376.
|
| [39] |
Broholm S K, Tähtiharju S, Laitinen R A, et al. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proceedings of the National Academy of Sciences of the United States of America, 2008,105(26):9117-9122.
|
 ), Zhao Xiaodong2, Zhen Pingping3, Chen Jing1, Chen Mingna1, Chen Na1, Pan Lijuan1, Wang Mian1, Xu Jing1, Yu Shanlin1, Chi Xiaoyuan1(
), Zhao Xiaodong2, Zhen Pingping3, Chen Jing1, Chen Mingna1, Chen Na1, Pan Lijuan1, Wang Mian1, Xu Jing1, Yu Shanlin1, Chi Xiaoyuan1( ), Zhang Jiancheng1(
), Zhang Jiancheng1( )
)