ASR(abscisic acid stress ripening)是从植物中发现的一类基因[7]编码的蛋白主要负责调控植物对干旱和盐胁迫的适应性、果实成熟、糖类代谢、低温、创伤、光感应以及激素的响应等,对维持植物的正常生命活动和生长发育发挥着重要作用。ASR基因是植物中特有的一类家族基因,ASR基因最早在番茄中通过干旱胁迫后分离克隆获得,之后在不同类型的植物中均有发现,但在拟南芥没有ASR基因存在,ASR家族成员最多的为玉米,有9个成员[8]。学者们对香蕉、荔枝、甜橙、桑树、葡萄、枇杷、甘蔗、茶树、火炬松、百合、木薯、番茄、大豆、水稻、玉米等植物的ASR基因进行了研究,其中,番茄ASR1基因的过表达能提高其对干旱和盐胁迫的耐受性[9],水稻ASR1基因在盐胁迫后茎中的表达量上调[10],百合ASR家族成员中LLA23的表达能提高百合对缺水和高盐胁迫的耐受性[11],火炬松ASR家族的Lp3基因也是参与水分亏缺调控的重要成员[12],海蓬子ASR1基因在盐胁迫下通过转录表达调控叶片中Na+的含量来提高植株的耐盐性[13]。
Feng等[14]对谷子ASR1基因的抗旱和耐氧化性进行了研究,结果显示,在拟南芥中过表达SiASR1基因能提高拟南芥植株对干旱和氧化胁迫的耐受性。李建锐[15]对谷子ASR4基因进行了克隆,并对ASR4与非生物胁迫的耐受性进行了相关研究分析,结果显示,SiASR4受脱落酸(abscisic acid,ABA)、PEG和盐的诱导表达。ASR基因的表达是一个极其复杂的过程,在不同组织部位ASR基因的转录表达量也有很大不同。本研究通过对谷子幼苗进行干旱和盐胁迫,取幼苗不同组织部位来分析ASR基因的表达,旨在分析谷子不同组织部位响应干旱和盐胁迫后ASR基因的表达情况,进而了解PEG、NaCl诱导后表达量较高的谷子ASR基因,为研究ASR基因在抗旱耐盐中的作用提供基础。
1 材料与方法
1.1 试验材料及培养方法
本试验于2018年8月在山西省农业科学院农作物品种资源研究所谷子糜子生理实验室进行。供试谷子品种豫谷1号为华北夏谷区的主栽品种,其具有抗病耐旱等优良特性。培养方式是使用有营养液的保湿水培盒置于光照培养箱中进行育苗,待幼苗长至4~5叶时,将幼苗的根部直接浸入10%的PEG-6000和150mmol/L的NaCl溶液中进行胁迫处理,分别取胁迫0、2、4、6、12、18h幼苗的根、茎、叶置于液氮中速冻,于-80℃冰箱中保存,用于提取RNA。
1.2 试验方法
1.2.1 谷子ASR家族基因的鉴定 在Phytozome v12.1的谷子数据库(Setaria italica v2.2)中下载谷子的蛋白序列文件,在数据库Protein family(Pfam)中通过关键词ABA/WDS查找谷子ASR家族蛋白的编号(PF02496),根据该蛋白的编号在谷子数据库中搜索下载所有ASR蛋白家族成员。之后通过NCBI中Batch CD search、Pfam的CD search来分析SiASRs是否具有ABA/WDS保守结构域,最终确定SiASRs家族基因数目。在NCBI谷子数据库下载谷子的染色体组数据,在JGI数据库确定8个谷子ASR家族成员的位置信息及其在染色体上的分布情况。并使用MapGene2Chromosome(MG2C)绘制SiASRs基因在染色体的分布图。
1.2.2 谷子ASR家族基因编码蛋白的理化特性分析
1.2.3 SiASRs蛋白家族系统进化树构建 下载水稻、高粱、玉米、大豆、番茄和柳枝稷中ASRs蛋白序列,运用Muscle对其ASR蛋白氨基酸序列和SiASRs蛋白氨基酸序列进行多重比对,用MEGA6.0分析比对结果,并采用邻接法(bootstrap replications of 1000)构建系统进化树[21]。
1.2.4 谷子ASR家族基因结构和启动子元件分析
在JGI官网下载谷子Setaria italica v2.2中的gff3文件,利用TBtools工具中的Redraw Gene Structure对谷子ASR家族基因的结构进行分析。通过MEME suit(http://meme-suite.org/)在线网站对SiASRs蛋白的motif元件进行搜索,分析各ASR蛋白的保守基序。利用Plantscare[22](http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)对SiASRs基因进行启动子分析。用Setaria italica v2.2下载谷子的全基因组序列和谷子gff3文件,使用TBtools将谷子全基因组序列提取出来,之后利用TBtools的Fasta Extractor将8个谷子ASR家族基因的序列全部提取出来,分析SiASRs基因编码区起始密码子ATG上游2 000bp基因组序列的启动子情况。
1.2.5 PEG、NaCl胁迫之后谷子ASR家族基因的表达分析 使用TIANGEN植物总RNA提取试剂盒和cDNA反转录试剂盒进行谷子总RNA的提取以及cDNA的反转录。对第一链cDNA进行PCR扩增,用1%琼脂糖凝胶进行电泳检测。用Premier 5.0进行荧光引物设计,利用Primer-BLAST对所设计引物进行特异性检测。用qRT-PCR中的相对定量法(2-△△Ct)分析8个ASR基因在逆境胁迫下的表达情况。
2 结果与分析
2.1 谷子ASR家族基因的鉴定及染色体定位分析
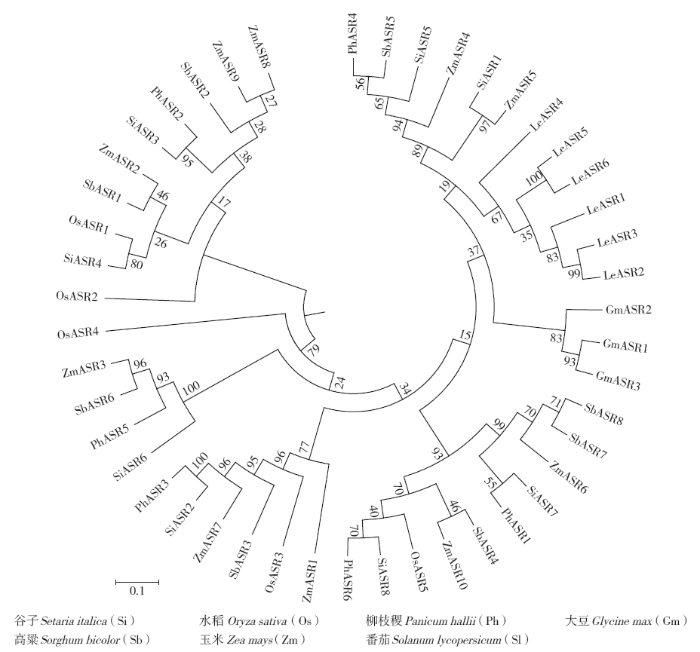
以ASR蛋白的保守结构域序列编号(PF02496)作为关键词,在JGI谷子数据库(Setaria italica v2.2)中进行搜索,结合NCBI中Batch CD search、Pfam中的CD search在谷子数据库中共搜索出了8个含有ABA/WDS完整结构域的ASR蛋白序列。MG2C工具绘制的SiASRs基因分布图(图1)显示,ASR1位于1号染色体,ASR2、ASR3、ASR4位于5号染色体,ASR5、ASR6、ASR7位于7号染色体,ASR8位于8号染色体。5和7号染色体上的ASR基因较多,各有3个SiASRs基因存在,1和8号染色体上各有1个ASR基因。谷子ASR基因属于小家族基因,在2、3、4、6、9号染色体上无ASR基因。
图1
2.2 谷子ASR家族基因编码蛋白的理化特性分析
SiASRs蛋白的等电点介于5.03~9.80,分子量介于11.48~26.00kDa,SiASR5蛋白的氨基酸数目最少,为101aa,SiASR6蛋白的氨基酸数目最多,为244aa。通过TMHMM分析SiASRs蛋白的跨膜结构,结果显示8个SiASRs蛋白均不存在跨膜结构。Protparam分析结果表明,8个SiASRs蛋白均为亲水性蛋白。8个SiASRs蛋白均无亲水性的吡咯赖氨酸(Pyl)和疏水性的硒半胱氨酸(Sec),只有SiASR2蛋白存在半胱氨酸(Cys),其余均缺乏半胱氨酸。
表1 谷子ASR家族基因成员及理化特性
Table 1
| 基因名称 Gene name | 基因ID Gene ID | 基因长度(bp) Length of gene | 编码区(bp)Length of CDS | 氨基酸数目(aa) Number of amino acids | 基因所在染色体的位置 Location of the genes on the chromosome | 等电点 PI | 分子量(kDa) Molecular weight | 亲水性指数 Hydrophilic index |
|---|---|---|---|---|---|---|---|---|
| SiASR1 | Seita.1G187200 | 876 | 324 | 107 | scaffold_1:26937730..26938605 reverse | 6.76 | 11.82 | -1.190 |
| SiASR2 | Seita.5G463600 | 1 020 | 522 | 173 | scaffold_5:46902215..46903234 forward | 6.26 | 19.38 | -1.277 |
| SiASR3 | Seita.5G463700 | 1 167 | 318 | 105 | scaffold_5:46906469..46907635 reverse | 9.73 | 11.71 | -1.147 |
| SiASR4 | Seita.5G463800 | 590 | 309 | 103 | scaffold_5:46907284..46907873 forward | 9.80 | 11.50 | -1.417 |
| SiASR5 | Seita.7G097100 | 803 | 306 | 101 | scaffold_7:19994321..19995123 reverse | 6.81 | 11.48 | -1.366 |
| SiASR6 | Seita.7G097200 | 1 318 | 735 | 244 | scaffold_7:19998219..19999536 reverse | 5.03 | 26.00 | -1.636 |
| SiASR7 | Seita.7G291500 | 984 | 417 | 138 | scaffold_7:33499331..33500314 forward | 5.88 | 15.45 | -1.298 |
| SiASR8 | Seita.8G045700 | 917 | 414 | 137 | scaffold_8:3645891..3646807 reverse | 6.15 | 15.42 | -1.346 |
2.3 ASRs 蛋白系统进化分析
利用MEGA6.0对水稻、玉米、高粱、大豆、番茄、柳枝稷和谷子的ASR蛋白进行多重序列比对后,采用邻接法(bootstrap replications of 1000)构建系统进化树(图2),从进化发育树可看出谷子ASR家族蛋白分为6大类,SiASR1和ZmASR5聚为一类,SiASR5和PhASR4、SbASR5进化关系较近,SiASR4和OsASR1、SiASR3和PhASR2分别聚为一个进化分支,SiASR2和PhASR3及SiASR6和ZmASR3、SbASR6、PhASR5聚为一类,SiASR7和PhASR1聚为一类,SiASR8和PhASR6进化关系最近。而大豆和番茄都为双子叶植物,在进化发育上都各自分为一类,且与单子叶植物的ASR蛋白在进化中差异很大。SiASRs蛋白与玉米ASR蛋白进化关系较近的有ZmASR3、ZmASR4、ZmASR5、ZmASR6;与水稻ASR蛋白进化关系较近的只有OsASR1和OsASR5;与高粱ASR蛋白进化关系较近的有SbASR1、SbASR5、SbASR6;与柳枝稷ASR蛋白进化关系较近的有PhASR1、PhASR2、PhASR3、PhASR4、PhASR5、PhASR6。表明谷子与柳枝稷的祖先亲缘关系更为相近。
图2
2.4 SiASRs家族基因结构和启动子顺式作用元件分布
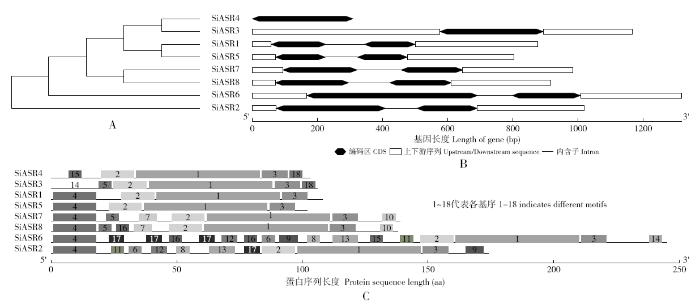
由结合SiASRs基因的编码区(CDS)序列和基因序列绘制的谷子ASR家族基因结构图(图3)可以看出,只有SiASR3、SiASR4没有内含子,SiASR1、SiASR2、SiASR5、SiASR6、SiASR7和SiASR8都有1个内含子。SiASR1和SiASR5、SiASR7和SiASR8在结构上非常相似,都为串联重复基因,彼此在进化上聚为一类。通过MEME网站鉴定了SiASRs蛋白的保守基序,共发现了18个基序元件。这些保守基序的长度短的有6个氨基酸,长的最多达到50个氨基酸。表2中列出了18个基序的详细信息。通过对Pfam、CDD的分析,motif1、motif2、motif3完全对应SiASRs蛋白的保守域(ABA/WDS)。SiASR1和SiASR5蛋白所含的基序都相同,都含有4个基序;SiASR3有6个基序响应元件;SiASR4有5个基序;SiASR7有7个基序响应元件,SiASR8比SiASR7多1个motif16元件,其余的响应元件均相同;SiASR2有11个基序响应元件;SiASR6蛋白拥有的基序数量最多。SiASR1、SiASR2、SiASR5、SiASR6、SiASR7和SiASR8蛋白都含有motif4。这与系统发育树的分类结果一致。
图3
图3
SiASRs蛋白系统进化树、SiASRs基因结构及SiASRs蛋白的基序分布
A,SiASRs蛋白的系统发育树;B,SiASRs基因结构图;C,SiASRs蛋白的保守基序
Fig.3
SiASRs protein phylogenetic tree, SiASRs gene structure and motif distribution of SiASRs protein
A, The phylogenetic tree of SiASRs protein; B, SiASRs gene structure map; C, The conserved motifs of SiASRs protein
表2 SiASRs蛋白的基序
Table 2
| 基序名称 Motif name | 基序宽度 Motif width | 基序的氨基酸组成 Amino acid composition of the motif |
|---|---|---|
| motif1 | 50 | QLGELGAVAAGAYALYEKHKAKKDPEHAHRHKIKEEVAAAAAVGSGGYAF |
| motif2 | 14 | EDYKKEEKEHKHKE |
| motif3 | 11 | HEHHEKKEAKK |
| motif4 | 18 | MAEEKHHHHYFHHHKDED |
| motif5 | 6 | QQPAGG |
| motif6 | 6 | SGTDEC |
| motif7 | 8 | TVAEEVVT |
| motif8 | 6 | DDCYNG |
| motif9 | 8 | RVGAGGYC |
| motif10 | 6 | KHHHLF |
| motif11 | 6 | HGSRRD |
| motif12 | 7 | VGRRGGG |
| motif13 | 11 | RNRAVGDDEYN |
| motif14 | 6 | KKHHFF |
| motif15 | 6 | DDEKNK |
| motif16 | 6 | YGGGYN |
| motif17 | 7 | YGRGGGD |
| motif18 | 6 | HRRHGH |
2.5 谷子ASR基因家族的启动子分析
通过TBtools中gff3 Sequeneces Extracter将谷子ASR基因编码区起始密码子ATG上游2 000bp的基因组序列提取出来,然后提交给PlantCARE Search for CARE。8个ASR基因涉及的响应元件包括生长素、茉莉酸甲酯(methyl jasmonate,MeJA)、ABA、赤霉素(gibberellin,GA)、水杨酸(salicylic acid,SA)、光、胁迫、低温、机械损伤等不同类型的响应元件。8个ASR基因都具有MeJa响应元件和光响应元件(G-Box);除ASR2基因外,其余7个ASR基因都有ABA响应元件(ABRE);ASR1和ASR6有低温响应元件(low temperature response element,LTR);ASR1、ASR5、ASR7和ASR8具有赤霉素响应元件(GARE);只有ASR4的启动子中存在防御和胁迫反应元件(TC-rich repeats);ASR2的启动子中具有机械损伤元件(WUN-motif)。ASR3启动子序列中的基序主要是光反应元件。
2.6 谷子ASR家族基因的表达分析
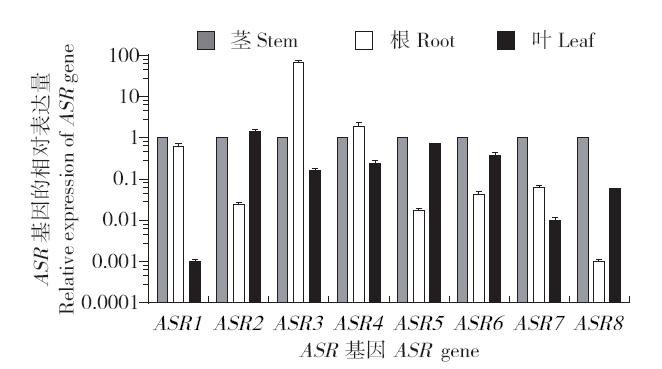
2.6.1 不同器官中ASR家族基因的表达分析 谷子不同组织部位的ASR基因表达分析结果:ASR1、ASR5、ASR6、ASR7和ASR8主要在茎中表达;ASR3、ASR4主要在根中表达;而ASR2主要在叶中表达(图4)。ASR3不同组织部位的相对表达量差异最大,其在根中的表达量为茎和叶中表达量的70倍左右,茎中的表达量很少。ASR2、ASR5、ASR6、ASR7和ASR8在根中的表达量较少,ASR1、ASR3、ASR5和ASR8在叶中的表达量相对于根部和茎部表达量非常少。因此,SiASRs基因表达具有较为明显的组织特异性。
图4
图4
胁迫前谷子ASR家族基因在根、茎、叶中的相对表达量
Fig.4
Relative expression of SiASR family genes in roots, stems and leaves before stress
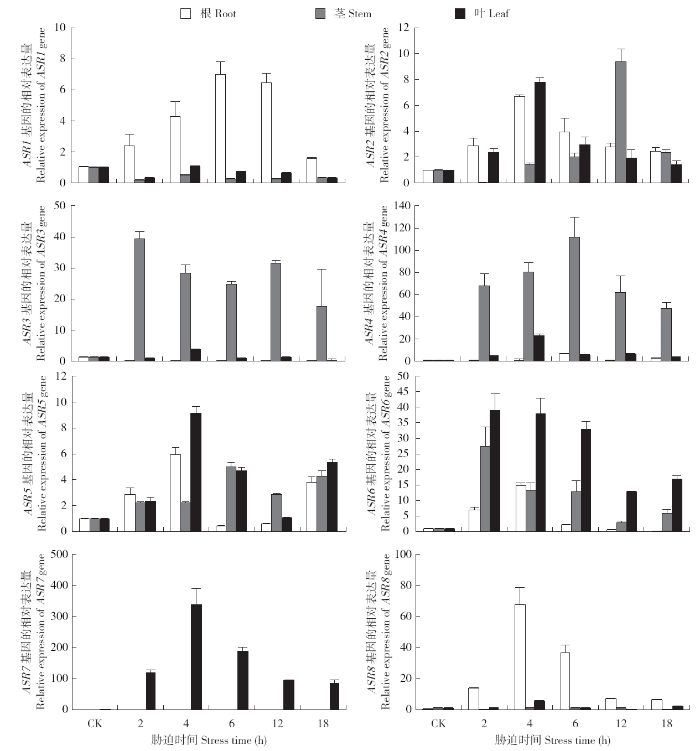
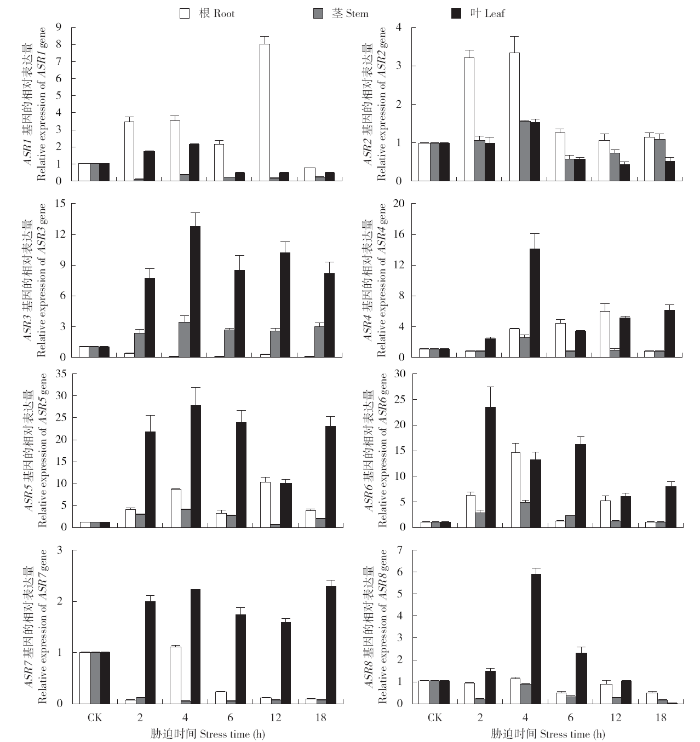
2.6.2 PEG-6000和NaCl胁迫后谷子ASR家族基因在根、茎和叶部的表达分析 在10%PEG-6000处理幼苗后,以谷子的18S rRNA基因作为内参,通过qRT-PCR的相对定量法对胁迫后8个ASR基因的相对表达量进行计算分析。从图5可以看出,ASR1总体表达量为根>叶>茎,ASR1在根部的表达于胁迫6h时达到峰值,为CK的7倍左右;而在茎部于胁迫4h时达峰值,表达量波动幅度不太明显;在叶中表达趋势与茎部类似,表达呈下调趋势。ASR2总体表达量为茎>叶>根,ASR2在10%PEG-6000处理后,根部的表达于胁迫4h时达峰值,为CK的6.5~7.0倍;随着胁迫时间的延长,茎部ASR2的表达先逐渐升高后降低,在胁迫12h时达到表达峰值,表达量大约为CK的9倍左右;在叶中的表达量于胁迫4h时达峰值,大约为CK的8倍左右。
图5
图5
10%PEG-6000胁迫处理后SiASR家族基因在根、茎和叶中的相对表达量
Fig.5
Relative expression of SiASR family genes in roots, stems and leaves after 10% PEG-6000 stress treatment
ASR3总体表达量为茎>叶>根,ASR3在根部的表达量逐渐下降;在茎部表达量呈急剧上升的状态,胁迫2h时表达量就达到CK的40倍左右,之后缓慢下降,在胁迫12h时又出现一个表达高峰,表达量为CK的30倍;在叶中ASR3的表达高峰于胁迫4h时达到CK的3.5倍左右。ASR4总体表达量为茎>叶>根,ASR4在根部的表达量变化相对于在茎部和叶中的变化波动较小,于胁迫6h时达到峰值为CK的5倍左右;在茎部的上调表达较为显著,胁迫2h时表达量就达到CK的70倍,在胁迫6h时表达量达到CK的110倍的顶峰;叶中则是先缓慢升高,之后缓慢降低,胁迫4h时达到CK表达量的20倍。
ASR5总体表达量为叶>根>茎,ASR5在叶中的表达情况为先升高,于胁迫4h时达到第1个峰值,胁迫6h时表达量恢复到CK表达量以下,胁迫18h时表达量又攀升到CK的4倍;茎部于胁迫6h时表达量最高,之后下降,胁迫12h时表达量又开始上升;在胁迫处理4h内叶中的表达情况与根部的表达趋势一致,胁迫12h时表达量恢复到CK表达量,胁迫18h时表达量又攀升到CK的6倍。ASR6总体表达量为叶>茎>根,ASR6在根中胁迫4h时表达量最高,胁迫12h时表达量几乎没有;茎部胁迫2h时表达量升高至CK的28倍左右,胁迫4~6h的表达量稳定,维持在CK的13倍左右;叶中ASR6的表达量,在胁迫2~6h这个时段内表达量维持在CK的30倍以上,之后略有降低,但总体叶中的表达量处于增加状态。
ASR7总体表达量为叶>茎>根,10%PEG-6000胁迫处理之后,ASR7在根部和茎部的表达量几乎未发生变化;但在叶中,胁迫2h时就升高至CK的100倍以上,胁迫4h时表达量达到CK的300倍以上,胁迫6h时开始逐渐降低。ASR8总体表达量为根>叶>茎,10%PEG-6000胁迫处理之后,ASR8在根中的表达量开始逐渐上升,胁迫4h时达到CK的65倍以上,之后开始下降;但ASR8在茎中的表达变化不明显;在叶中只在胁迫4h时出现1个表达峰值。
综上所述,谷子ASR家族基因对干旱胁迫具有非常积极的响应,ASR3、ASR4在茎部的上调表达量为CK的数十倍,ASR6在茎叶中的上调表达幅度大,ASR7在叶中上调表达达到CK的300倍以上,ASR8在根部的响应表达最明显。且ASR7和ASR8达到表达峰值的时间都为胁迫处理4h时。
从图6可看出,经过150mmol/L NaCl溶液胁迫,谷子ASR家族基因在感受到盐胁迫之后,所有的ASR基因在叶中都呈上调表达,ASR5、ASR6在叶中的上调表达最为突出。在茎部ASR2、ASR3、ASR4、ASR5、ASR6的表达量略微上调,都维持在CK的2~4倍;而ASR1、ASR7、ASR8在盐胁迫之后都开始下调表达。在根部ASR1、ASR2、ASR4、ASR5、ASR6的表达都为上调,ASR3、ASR7和ASR8在根部为下调表达。因此,谷子的ASR家族基因在盐胁迫之后,都参与了代谢过程,以维持植物正常的生理活动。
图6
图6
盐胁迫后根、茎、叶中SiASR家族基因的相对表达量
Fig.6
Relative expression of SiASR family genes in roots, stems and leaves after salt stress
盐胁迫后,ASR1总体表达量为根>叶>茎,盐胁迫后,ASR1基因在根和叶中的表达量开始逐渐增加,分别于胁迫12h时和胁迫4h时达到峰值。盐胁迫后,ASR2总体表达量为根>茎>叶,ASR2基因在根、茎、叶中的表达趋势一致,都在胁迫4h时达到最大表达量,在胁迫4h时根中的表达量达到CK的3倍。盐胁迫后,ASR3总体表达量为叶>根>茎,ASR3对该逆境发生了响应,在根部的表达量下调;在茎部的表达量趋于稳定;在叶中的表达也是从胁迫2h时开始逐渐上升,在胁迫4h时达到CK的13倍左右,之后一直维持在10倍左右。ASR4在根、茎、叶的表达都是上调表达,ASR4总体表达量为叶>根>茎。根部在感应到胁迫之后开始上调表达,胁迫12h时达到CK的7倍;在叶中的表达变化最明显,胁迫4h时表达量为CK的13倍,茎部的表达不明显。ASR5对盐胁迫的响应最敏感,ASR5总体表达量为叶>根>茎,在叶中的上调表达明显,在胁迫4h时,表达量达到CK的25倍以上;而且在根部的响应也较明显,胁迫12h时表达量最高,是CK的10倍以上;随胁迫时间延长,在茎部的表达呈现先上调后下调的趋势。ASR6是对盐胁迫较敏感的基因,ASR6总体表达量为叶>根>茎,在根部接受到逆境信号后,表达量开始上调,在胁迫4h时达到CK的13倍左右;在叶中的上调表达最迅速,胁迫2h时上调至CK的23倍左右;茎部的表达先上调后下调,在胁迫4h时表达量达到峰值,为CK的5倍。盐胁迫之后,ASR7和ASR8总体表达量为叶>根>茎,ASR7基因在根和茎部都是下调表达,只有在叶中呈上调表达,但上调幅度较小。ASR8基因在根和茎部也都是下调表达,在叶中整体上调表达,胁迫4h时达到CK的6倍以上。
3 讨论
3.1 谷子ASR家族基因生物信息学分析
ASR家族基因是植物特有的一类转录因子家族基因,但不是所有的植物都存在ASR基因,双子叶植物的ASR蛋白与单子叶的ASR蛋白没有同源性。谷子拥有8个ASR基因,属于小家族基因,且编码的蛋白序列较短,大部分ASR蛋白都具有由70个左右的氨基酸残基构成的ABA/WDS保守结构域,ASR7和ASR8蛋白的保守结构域由50个左右的氨基酸残基组成。谷子8个ASR基因所包含的响应元件涉及植物对激素、光、胁迫等不同信号类型的响应元件。8个ASR基因都具有MeJA响应元件和G-Box光响应元件;MeJA是与损伤相关的植物激素和信号分子,因此ASR蛋白在植物体内的调控表达可能起防御和调节作用,以保护植物免受外界因素的伤害。谷子ASR基因启动子中广泛存在光响应元件和ABRE元件,ABRE元件与各类逆境调控相关,它可以调控ASR基因的表达,以提高植物本身的抗逆性。
3.2 谷子ASR基因在不同器官中的表达分析
不同器官中ASR基因的表达情况:ASR1的相对表达量为茎>根>叶,ASR2的相对表达量为叶>茎>根,ASR3主要在根部表达,在茎叶中几乎不表达。ASR4的相对表达量为根>茎>叶,ASR5主要在茎和叶中表达,根部几乎不表达。ASR6的相对表达量为茎>叶>根,ASR7的相对表达量为茎>根>叶,ASR8的相对表达量为茎>叶>根。且ASR3的相对表达量在8个基因中是最高的。可见SiASRs基因表达具有较为明显的组织特异性。
3.3 谷子ASR基因在PEG-6000胁迫下不同器官中的表达分析
10%PEG-6000胁迫下ASR1主要在根部转录表达;ASR2在根、茎、叶3个部位都呈上调表达;ASR3和ASR4主要在茎部表达,预测这2个基因可能在调控茎部水分的运输中起作用;ASR5和ASR6在根、茎、叶3个部位都呈上调表达;而ASR7主要在叶中表达,可能具有调节叶片气孔关闭的功能;ASR8主要在根部表达,可能在感受到干旱胁迫后,调节植物根部以增加根部的吸水量来维持植物正常的生命活动。李建锐[15]研究结果显示,在PEG胁迫处理后,ASR1和ASR3的表达量提升最高,ASR1在胁迫1h表达量就达到0h的310.12倍,ASR3在胁迫12h时表达量达到0h的1 957.78倍;本研究中,在干旱胁迫之后表达量最高的2个基因是ASR4和ASR7,ASR4在胁迫6h时茎部的表达量达到CK的110倍以上;ASR7主要在叶中表达,在胁迫4h时表达量达到CK的310倍左右。
3.4 谷子ASR基因在NaCl胁迫下不同器官中的表达分析
NaCl胁迫后,8个SiASRs基因在叶中上调表达都较为明显;根部响应盐胁迫后表达上调的SiASRs基因有5个(ASR1、ASR2、ASR4、ASR5和ASR6),下调表达的有ASR3和ASR7,ASR8在根部的表达较为稳定;在茎部下调表达的基因有ASR1、ASR7和ASR8,其余5个ASR基因都有略微上调。在NaCl处理幼苗后,表达量最高的2个基因是ASR5和ASR6,ASR5于胁迫4h时在叶中的表达量达到CK的27倍左右,ASR6于胁迫2h时达到最大表达量,约为CK的23倍左右。李建锐[15]研究结果为,幼苗在NaCl溶液处理后,谷子ASR3和ASR4基因的表达量与胁迫0h的表达量相比提高倍数最大,且都在胁迫9h时达到表达最高峰,分别是胁迫0h的192.01和85.63倍。
干旱和盐胁迫是植物非生物胁迫中最主要的两类胁迫,也是影响植物生长发育的主要因素,在作物生长发育的关键时期植株体内水分的过度缺失会影响其正常的代谢活动。ASR蛋白是调节植物渗透胁迫中重要的一类转录因子,Yang等[11]将百合ASR1基因转化到拟南芥中过量表达,转基因的拟南芥种子能在具有NaCl的溶液中萌发,转基因拟南芥的水分散失减少,抗干旱和抗盐能力增强。本研究中,谷子ASR基因的表达明显受干旱和盐胁迫的影响,证明谷子ASR基因家族是调控干旱和盐胁迫的重要家族基因。
4 结论
谷子ASR基因家族共有8个成员,且8个ASR基因编码的蛋白均为亲水性蛋白。除ASR2外,谷子ASR基因启动子中都存在ABRE元件,ABRE元件与各类逆境调控相关,可以提高植物本身的抗逆性。在谷子ASR基因家族中对干旱胁迫响应最强的2个基因为ASR4和ASR7,而对盐胁迫响应最强烈的2个基因为ASR5和ASR6,说明ASR基因家族的不同成员间应对非生物胁迫的功能也有所差异。
参考文献
谷子[Setaria italica (L.) P. Beauv.]作为功能基因组研究模式植物的发展现状及趋势
Tomato Asr1 mRNA and protein are transiently expressed following salt stress,osmotic stress and treatment with abscisic acid
DOI:10.1016/0168-9452(95)94515-K URL [本文引用: 1]
Rice ASR1 has function in abiotic stress tolerance during early growth stages of rice
DOI:10.1007/s13765-013-3060-6
URL
[本文引用: 1]

OsASR1 expression was induced through Abscisic acid (ABA) and stress treatments in leaves. The constitutive overexpression of OsASR1 in rice reduced ABA sensitivity, and increased high salinity and osmotic stress tolerance in early growth stages. These results indicated that OsASR1 has function in abiotic stress tolerance during early growth stages of rice.
A lily asr protein involves abscisic acid signaling and confers drought and salt resistance in Arabidopsis
DOI:10.1104/pp.105.065458
URL
PMID:16169963
[本文引用: 2]

LLA23, an abscisic acid-, stress-, and ripening-induced protein, was previously isolated from lily (Lilium longiflorum) pollen. The expression of LLA23 is induced under the application of abscisic acid (ABA), NaCl, or dehydration. To provide evidence on the biological role of LLA23 proteins against drought, we used an overexpression approach in Arabidopsis (Arabidopsis thaliana). Constitutive overexpression of LLA23 under the cauliflower mosaic virus 35S promoter confers reduced sensitivity to ABA in Arabidopsis seeds and, consequently, a reduced degree of seed dormancy. Transgenic 35SLLA23 seeds are able to germinate under unfavorable conditions, such as inhibitory concentrations of mannitol and NaCl. At the molecular level, altered expression of ABA/stress-regulated genes was observed. Thus, our results provide strong in vivo evidence that LLA23 mediates stress-responsive ABA signaling. In vegetative tissues, it is intriguing that Arabidopsis 35SLLA23 stomata remain opened upon drought, while transgenic plants have a decreased rate of water loss and exhibit enhanced drought and salt resistance. A dual function of the lily abscisic acid-, stress-, and ripening-induced protein molecule is discussed.
Analysis and localization of the Water-Deficit Stress-Induced gene (lp3)
DOI:10.1007/s00344-002-0128-7
URL
[本文引用: 1]

LP3 is a water-deficit-induced protein, which is highly homologous to ASR (ABA, stress and ripening) proteins. Homology was found in the C-terminal region of the putative LP3 protein while lower homologies were found in the N-terminal region. The goal of this study was to investigate the function of the LP3 protein and the mechanism of the lp3 promoter in response to water-deficit stress (WDS) and other stresses. In regenerated transgenic tobacco (T0), expression of β-glucuronidase (GUS) from the lp3 promoter-GUS construct was observed in polyethylene glycol (PEG), abscisic acid (ABA), methyl-jasmonate (MeJa), and fluridone (Flu) treatments. GUS expression was not observed following gibberellin (GA3), 2-methyl-4-dichlorophenoxy acetic acid (2,4-D), silver nitrate, or ethephon (ethylene releasing agent) treatments. Germinated T1 seedlings containing the lp3 promoter-GUS construct exhibited GUS activity up to 40 days postgermination. Expression could be restored when 5-azacytidine was included in the culture media, indicative of a developmentally regulated silencing mechanism involving methylation. In transgenic tobacco, the LP3 protein localized in the cell nucleus was induced by WDS and appeared to be developmentally regulated.
The SbASR-1 gene cloned from an extreme halophyte Salicornia brachiata enhances salt tolerance in transgenic tobacco
DOI:10.1007/s10126-012-9442-7
URL
[本文引用: 1]

Salinity severely affects plant growth and development. Plants evolved various mechanisms to cope up stress both at molecular and cellular levels. Halophytes have developed better mechanism to alleviate the salt stress than glycophytes, and therefore, it is advantageous to study the role of different genes from halophytes. Salicornia brachiata is an extreme halophyte, which grows luxuriantly in the salty marshes in the coastal areas. Earlier, we have isolated SbASR-1 (abscisic acid stress ripening-1) gene from S. brachiata using cDNA subtractive hybridisation library. ASR-1 genes are abscisic acid (ABA) responsive, whose expression level increases under abiotic stresses, injury, during fruit ripening and in pollen grains. The SbASR-1 transcript showed up-regulation under salt stress conditions. The SbASR-1 protein contains 202 amino acids of 21.01-kDa molecular mass and has 79 amino acid long signatures of ABA/WDS gene family. It has a maximum identity (73 %) with Solanum chilense ASR-1 protein. The SbASR-1 has a large number of disorder-promoting amino acids, which make it an intrinsically disordered protein. The SbASR-1 gene was over-expressed under CaMV 35S promoter in tobacco plant to study its physiological functions under salt stress. T-0 transgenic tobacco seeds showed better germination and seedling growth as compared to wild type (Wt) in a salt stress condition. In the leaf tissues of transgenic lines, Na+ and proline contents were significantly lower, as compared to Wt plant, under salt treatment, suggesting that transgenic plants are better adapted to salt stress.
Investigation of the ASR family in foxtail millet and the role of ASR1 in drought/oxidative stress tolerance
DOI:10.1007/s00299-015-1873-y
URL
PMID:26441057
[本文引用: 1]

Six foxtail millet ASR genes were regulated by various stress-related signals. Overexpression of ASR1 increased drought and oxidative tolerance by controlling ROS homeostasis and regulating oxidation-related genes in tobacco plants. Abscisic acid stress ripening (ASR) proteins with ABA/WDS domains constituted a class of plant-specific transcription factors, playing important roles in plant development, growth and abiotic stress responses. However, only a few ASRs genes have been characterized in crop plants and none was reported so far in foxtail millet (Setaria italic), an important drought-tolerant crop and model bioenergy grain crop. In the present study, we identified six foxtail millet ASR genes. Gene structure, protein alignments and phylogenetic relationships were analyzed. Transcript expression patterns of ASR genes revealed that ASRs might play important roles in stress-related signaling and abiotic stress responses in diverse tissues in foxtail millet. Subcellular localization assays showed that SiASR1 localized in the nucleus. Overexpression of SiASR1 in tobacco remarkably increased tolerance to drought and oxidative stresses, as determined through developmental and physiological analyses of germination rate, root growth, survival rate, relative water content, ion leakage, chlorophyll content and antioxidant enzyme activities. Furthermore, expression of SiASR1 modulated the transcript levels of oxidation-related genes, including NtSOD, NtAPX, NtCAT, NtRbohA and NtRbohB, under drought and oxidative stress conditions. These results provide a foundation for evolutionary and functional characterization of the ASR gene family in foxtail millet.
Shotgun proteomics and network analysis between plasma membrane and extracellular matrix proteins from rat olfactory ensheathing cells
DOI:10.3727/096368910X492607
URL
PMID:20350363
[本文引用: 1]

Olfactory ensheathing cells (OECs) are a special type of glial cells that have characteristics of both astrocytes and Schwann cells. Evidence suggests that the regenerative capacity of OECs is induced by soluble, secreted factors that influence their microenvironment. These factors may regulate OECs self-renewal and/or induce their capacity to augment spinal cord regeneration. Profiling of plasma membrane and extracellular matrix through a high-throughput expression proteomics approach was undertaken to identify plasma membrane and extracellular matrix proteins of OECs under serum-free conditions. 1D-shotgun proteomics followed with gene ontology (GO) analysis was used to screen proteins from primary culture rat OECs. Four hundred and seventy nonredundant plasma membrane proteins and 168 extracellular matrix proteins were identified, the majority of which were never before reported to be produced by OECs. Furthermore, plasma membrane and extracellular proteins were classified based on their protein-protein interaction predicted by STRING quantitatively integrates interaction data. The proteomic profiling of the OECs plasma membrane proteins and their connection with the secretome in serum-free culture conditions provides new insights into the nature of their in vivo microenvironmental niche. Proteomic analysis for the discovery of clinical biomarkers of OECs mechanism warrants further study.
Genome-wide identification and expression analysis of the expansin gene family in tomato
DOI:10.1007/s00438-015-1133-4
URL
PMID:26499956
[本文引用: 1]

Plant expansins are capable of inducing pH-dependent cell wall extension and stress relaxation. They may be useful as targets for crop improvement to enhance fruit development and stress resistance. Tomato is a major agricultural crop and a model plant for studying fruit development. Because only some tomato expansins have been studied, a genome-wide analysis of the tomato expansin family is necessary. In this study, we identified 25 SlEXPAs, eight SlEXPBs, one SlEXLA, four SlEXLBs, and five short homologs in the tomato genome. 25 of these genes were identified as being expressed. Bioinformatic analysis showed that although tomato expansins share similarities with those from other plants, they also exhibit specific features regarding genetic structure and amino acid sequences, which indicates a unique evolutionary process. Segmental and tandem duplication events have played important roles in expanding the tomato expansin family. Additionally, the 3-exon/2-intron structure may form the basic organization of expansin genes. We identified new expansin genes preferentially expressed in fruits (SlEXPA8, SlEXPB8, and SlEXLB1), roots (SlEXPA9, SlEXLB2, and SlEXLB4), and floral organs. Among the analyzed genes those that were inducible by hormone or stress treatments, including SlEXPA3, SlEXPA7, SlEXPB1-B2, SlEXPB8, SlEXLB1-LB2, and SlEXLB4. Our findings may further clarify the biological activities of tomato expansins, especially those related to fruit development and stress resistance, and contribute to the genetic modification of tomato plants to improve crop quality and yield.
Plant CARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences
DOI:10.1093/nar/30.1.325
URL
PMID:11752327
[本文引用: 1]

PlantCARE is a database of plant cis-acting regulatory elements, enhancers and repressors. Regulatory elements are represented by positional matrices, consensus sequences and individual sites on particular promoter sequences. Links to the EMBL, TRANSFAC and MEDLINE databases are provided when available. Data about the transcription sites are extracted mainly from the literature, supplemented with an increasing number of in silico predicted data. Apart from a general description for specific transcription factor sites, levels of confidence for the experimental evidence, functional information and the position on the promoter are given as well. New features have been implemented to search for plant cis-acting regulatory elements in a query sequence. Furthermore, links are now provided to a new clustering and motif search method to investigate clusters of co-expressed genes. New regulatory elements can be sent automatically and will be added to the database after curation. The PlantCARE relational database is available via the World Wide Web at http://sphinx.rug.ac.be:8080/PlantCARE/.
甘蓝型油菜PIN家族基因的鉴定与生物信息学分析
DOI:10.3724/SP.J.1006.2018.01334
URL
[本文引用: 1]

PIN家族基因是一类调控植物生长素极性运输的重要载体元件, PIN基因编码生长素输出蛋白, 介导生长素在植物体的运输, 然而在基因组较复杂的甘蓝型油菜中缺乏系统研究。本研究运用生物信息学方法在甘蓝型油菜全基因组数据库筛选甘蓝型油菜PIN家族基因, 对鉴定出的29个BnPINs基因开展拷贝数变异、分子特征、跨膜结构域、保守基序、染色体定位、系统进化树构建、PIN蛋白二级结构及三级结构预测等研究, 结合高通量转录组测序进行低氮胁迫下的转录水平分析。结果表明, 甘蓝型油菜PIN家族基因拷贝数明显多于拟南芥、甘蓝和白菜所具有的PIN家族基因数量; BnPINs蛋白多属于由碱性氨基酸组成的稳定蛋白, 含有保守的N末端结构域, 二级结构与拟南芥PIN蛋白相似; 系统进化选择能力分析表明, BnPINs基因与甘蓝和白菜PIN家族基因进化关系相近。转录组测序表明, BnPIN1s、BnPIN2s、BnPIN3s基因主要在甘蓝型油菜根部表达且受长期低氮(72 h)诱导, BnPIN6s和BnPIN8s基因主要在地上部表达, 低氮会抑制BnPIN6s表达。本研究结果为进一步研究甘蓝型油菜PIN家族基因生物学功能尤其是在响应低氮胁迫中的功能奠定基础, 为已知大量数据的其他物种家族基因生物信息学研究提供参考。