水稻抽穗期的遗传基础复杂,属于数量性状。目前发掘的水稻抽穗期相关的QTL已超过700个,位于第3号染色体的QTL数目最多,但目前仅有29个QTL被克隆[5]。Kojima等[6]将水稻抽穗期相关QTL精细定位到20kb的范围内,并在该区间克隆到1个与水稻抽穗期相关的基因Hd3a。Zong等[7]研究发现,在长日照或短日照下,敲除Ghd7及DTH8会促使水稻抽穗,在长日照条件下,Hd1会促使Ghd7表达,最终导致水稻抽穗期推迟甚至不抽穗。Li等[8]发现,HGW突变体植株的抽穗期延迟,这一基因编码泛素相关的结构蛋白。尽管目前定位到的QTL数量很多,但大多数QTL的区间很大,且水稻抽穗期属于复杂的数量性状,因此,需要进一步挖掘新的QTL。
本研究利用空育131和小白粳子衍生的重组自交系群体(RIL),通过靶向测序基因型检测技术(genotyping by target sequencing,GBTS)进行基因型分析,构建了包含527个bin标记的高密度遗传图谱,发掘新的抽穗期相关主效QTL,为水稻基因克隆及分子标记辅助选择育种提供参考。
1 材料与方法
1.1 试验材料
以粳稻品种空育131为母本、陆稻品种小白粳子为父本配制杂交组合,以通过“单粒传”法得到的包含195个株系的F9群体为试验材料。
1.2 试验方法
所有材料种植于东北农业大学阿城水稻试验基地,每份材料种植8行,每行20株,行距30cm、株距13.3cm,均为单株插秧。按株系单独编号,调查并记录抽穗日期。当该株系的第1个稻穗尖露出剑叶叶鞘至少1cm时,记为该株系的抽穗日期,每隔1d调查1次,将株系从播种到抽穗的天数作为该株系的抽穗期表型值,方法参照文献[11],略有改动。田间管理依照常规大田生产。
1.3 数据统计分析
使用Excel 2016和SPSS Statistics 19.0进行相关性状表型数据统计分析。
1.4 遗传连锁图谱的构建及QTL分析
利用10K SNP芯片(石家庄博瑞迪生物技术有限公司)的靶向测序基因型检测技术进行RIL群体的基因型分析,经由双亲多态性分析及IciMapping-Bin模块的去冗余分析,将RIL群体最小重组区间内的多个SNP标记合并为1个“bin”,最终获得527个高质量多态性bin标记。通过JoinMap 4.0进行遗传连锁图谱的构建,该图谱共覆盖水稻基因组1874.85cM,标记间的平均遗传距离为3.56cM。采用QTL IciMapping 4.2软件的完备区间作图法(ICIM)进行QTL定位(
2 结果与分析
2.1 水稻抽穗期的表型分析
分别对亲本及RIL群体的抽穗期进行基本统计分析(表1)。亲本的抽穗期处于RIL群体范围内,抽穗期在双亲之间表现出极显著差异,小白粳子的抽穗期明显长于空育131。RIL群体的抽穗期具有超亲分离现象,平均为97.74d,变异范围为76.00~111.00d,变异系数为7.68%。通过数据正态分布的适合性检验后发现,抽穗期的偏度和峰度绝对值均小于1,说明性状的表型符合正态分布,具有典型的数量性状遗传特征。
表1 水稻抽穗期的基本统计分析
Table 1
| 项目 Item | 亲本Parents | 重组自交系群体RIL population | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 空育131 Kongyu 131 (d) | 小白粳子 Xiaobaijingzi (d) | 平均值 Mean (d) | 标准差 Standard deviation | 变异范围 Range (d) | 变异系数 Coefficient of variation (%) | 峰度 Kurtosis | 偏度 Skewness | ||
| 抽穗期Heading date | 91.00 | 104.00** | 97.74 | 7.51 | 76.00~111.00 | 7.68 | -0.42 | -0.23 | |
“**”表示在0.01水平上极显著差异
“**”indicates extremely significant difference at 0.01 level
2.2 水稻抽穗期QTL分析
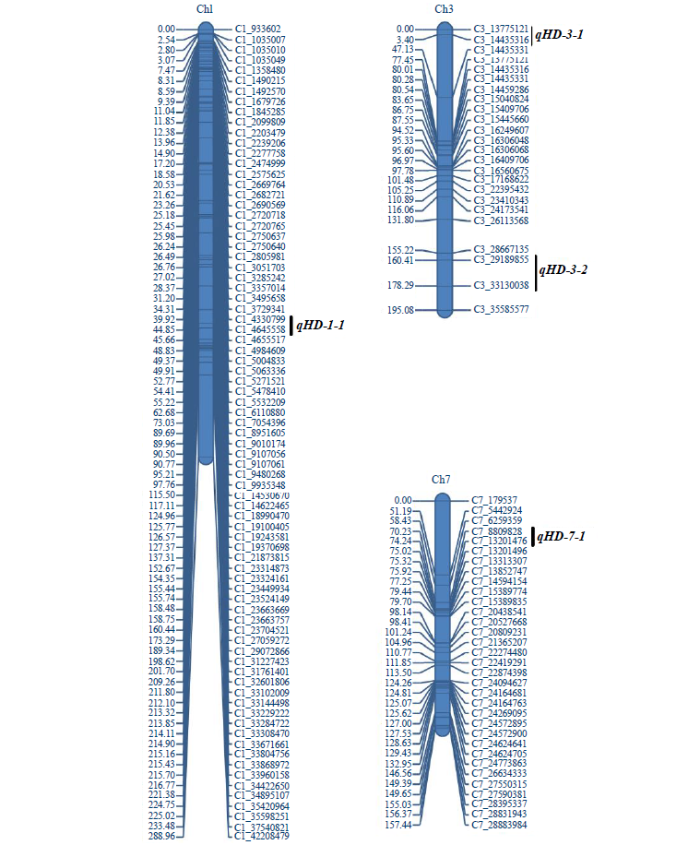
采用IciMapping 4.2软件的完备区间作图法,对水稻抽穗期进行QTL分析(图1和表2),在水稻第1、3和7号染色体上共检测到4个与水稻抽穗期相关的QTL。4个QTL的LOD值在3.0181~13.2288之间,表型贡献率介于4.0619%~21.7016%。其中,qHD-3-1和qHD-7-1的表型贡献率均大于10%,属于主效QTL。除qHD-7-1的增效等位基因来自于小白粳子外,其余3个QTL的增效等位基因均来自于空育131。通过与已克隆水稻抽穗期相关基因比较发现,qHD-1-1、qHD-3-2和qHD-7-1区间内分别含有已知基因OsGI、Hd6和Ghd7。而qHD-3-1区间内未发现已克隆基因,是控制水稻抽穗期的新位点。qHD-3-1贡献率为10.5098%,位于标记C3_799346和C3_1493308之间,物理距离为0.69Mb。
图1
图1
水稻抽穗期QTL在染色体上的分布
Fig.1
Distribution of QTLs on chromosomes at heading stage in rice
表2 水稻抽穗期QTL及其遗传效应
Table 2
| 数量性状位点 QTL | 染色体 Chromosome | 标记区间 Marker interval | LOD值 LOD peak | 贡献率 Contribution rate (%) | 加性效应 Additive effect | 已知基因 Known gene | |
|---|---|---|---|---|---|---|---|
| qHD-1-1 | 1 | C1_4330799 | C1_4645558 | 3.0181 | 4.0619 | 1.5969 | OsGI |
| qHD-3-1 | 3 | C3_799346 | C3_1493308 | 7.0671 | 10.5098 | 2.7637 | |
| qHD-3-2 | 3 | C3_29189855 | C3_33130038 | 3.2297 | 4.5338 | 1.6866 | Hd6 |
| qHD-7-1 | 7 | C7_8809828 | C7_13201476 | 13.2288 | 21.7016 | -3.8409 | Ghd7 |
3 讨论
水稻抽穗期与其产量、品质及适应性密切相关[14]。但传统的遗传标记(例如SSR、RFLP等)存在密度低、数量少及精度不足甚至双交换位点漏测等问题,影响了抽穗期相关QTL的发掘[15]。相比之下,通过bin标记构建的高密度遗传图谱省时省力、标记密度高且能够直接发掘区间内的候选基因,并且由于单个bin内部包含多个不重组的SNP,避免了双交换位点漏测的问题。本研究利用包含527个bin标记的高密度遗传图谱,平均遗传距离仅为3.56cM,能够快速高效地发掘抽穗期相关QTL,并进一步预测候选基因。通过对由空育131和小白粳子衍生的RIL群体进行抽穗期调查,发现水稻抽穗期在双亲中表现出极显著差异,RIL群体变异范围较广,有明显的超亲分离现象,表型符合正态分布,具有典型的数量性状遗传特征,适于进行QTL分析。
本研究所定位的4个与抽穗期相关QTL中,其中3个含有与抽穗期相关的基因。Izawa等[16]发现,Os-GI突变体在短日照下表现为晚花;Takahashi等[17]研究表明,来源于亲本Kasalath的Hd6等位基因在长日照及自然生长条件下均延迟抽穗,反之,在短日照下则不能延迟抽穗。Xue等[18]发现,长日照条件提高Ghd7的表达,能推迟抽穗期。因此,Os-GI、Hd6和Ghd7能够对提高全球水稻产量及水稻季节适应性起到重要作用。此外,只有位于3号染色体上的qHD-3-1中未包含报道过的已知基因,且其贡献率高达10.5098%,是新的抽穗期相关QTL,本结果能够为水稻抽穗期QTL的精细定位及克隆提供参考,同时为分子标记辅助育种提供了新的资源。
4 结论
基于GBTS技术对双亲空育131、小白粳子及其衍生的RIL群体进行基因型分析,构建了包含527个bin标记的高密度遗传图谱,通过QTL分析,鉴定到4个与水稻抽穗期相关的QTL。其中除qHD-3-1外,其余3个与抽穗期相关QTL区间内均包含已知的抽穗期基因,说明qHD-3-1是控制水稻抽穗期的新位点。
参考文献
Reinventing rice to feed the world
DOI:10.1126/science.321.5887.330 PMID:18635770 [本文引用: 1]
Mapping of quantitative trait loci controlling heading date among rice cultivars in the northern most region of Japan
DOI:10.1270/jsbbs.58.367 URL [本文引用: 1]
Identification of stably expressed QTL for heading date using reciprocal introgression line and recombinant inbred line populations in rice
DOI:10.1017/S0016672312000444 URL [本文引用: 1]
Hd3a,a rice ortholog of the Arabidopsis FT gene,promotes transition to flowering downstream of Hd1 under short-day conditions
Heading date 3a (Hd3a) has been detected as a heading-date-related quantitative trait locus in a cross between rice cultivars Nipponbare and Kasalath. A previous study revealed that the Kasalath allele of Hd3a promotes heading under short-day (SD) conditions. High-resolution linkage mapping located the Hd3a locus in a approximately 20-kb genomic region. In this region, we found a candidate gene that shows high similarity to the FLOWERING LOCUS T (FT) gene, which promotes flowering in Arabidopsis: Introduction of the gene caused an early-heading phenotype in rice. The transcript levels of Hd3a were increased under SD conditions. The rice Heading date 1 (Hd1) gene, a homolog of CONSTANS (CO), has been shown to promote heading under SD conditions. By expression analysis, we showed that the amount of Hd3a mRNA is up-regulated by Hd1 under SD conditions, suggesting that Hd3a promotes heading under the control of Hd1. These results indicate that Hd3a encodes a protein closely related to Arabidopsis FT and that the function and regulatory relationship with Hd1 and CO, respectively, of Hd3a and FT are conserved between rice (an SD plant) and Arabidopsis (a long-day plant).
Strong photoperiod sensitivity is controlled by cooperation and competition among Hd1,Ghd7 and DTH8 in rice heading
DOI:10.1111/nph.v229.3 URL [本文引用: 1]
The rice HGW gene encodes a ubiquitin-associated (UBA) domain protein that regulates heading date and grain weight
DOI:10.1371/journal.pone.0034231 URL [本文引用: 1]
Fine-mapping quantitative trait loci for body weight and abdominal fat traits:effects of marker density and sample size
DOI:10.3382/ps.2007-00512
PMID:18577610
[本文引用: 1]

Highly significant QTL for BW and abdominal fat traits on chicken chromosome 1 were reported previously in a unique F2 population. The objective of this study was to confirm and refine the QTL locations. Compared with the previous experiment, this study added 8 new families, including all the animals in the pedigree, and genotyped 9 more microsatellite markers, including 6 novel ones. Linkage analyses were performed. The results of the linkage analyses showed that the confidence intervals for BW and abdominal fat percentage were narrowed sharply to a small interval spanning 5.5 and 3.7 Mb, respectively. The results of the present study showed that using more markers and individuals could decrease the confidence interval of QTL effectively. In the current QTL region, by combining the biological knowledge of genes and the results of a microarray analysis that was performed in divergently selected lean and fat lines, several genes stood out as potential candidate genes.
High-throughput SNP genotyping to accelerate crop improvement
DOI:10.9787/PBB.2014.2.3.195 URL [本文引用: 1]
Empirical threshold values for quantitative trait mapping
The detection of genes that control quantitative characters is a problem of great interest to the genetic mapping community. Methods for locating these quantitative trait loci (QTL) relative to maps of genetic markers are now widely used. This paper addresses an issue common to all QTL mapping methods, that of determining an appropriate threshold value for declaring significant QTL effects. An empirical method is described, based on the concept of a permutation test, for estimating threshold values that are tailored to the experimental data at hand. The method is demonstrated using two real data sets derived from F(2) and recombinant inbred plant populations. An example using simulated data from a backcross design illustrates the effect of marker density on threshold values.
Gene nomenclature system for rice
DOI:10.1007/s12284-008-9004-9 URL [本文引用: 1]
A whole‐genome SNP array (RICE 6 K) for genomic breeding in rice
DOI:10.1111/pbi.2013.12.issue-1 URL [本文引用: 1]
Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field
DOI:10.1105/tpc.111.083238 URL [本文引用: 1]
Hd6,a rice quantitative trait locus involved in photoperiod sensitivity,encodes the α subunit of protein kinase CK2
Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice
DOI:10.1038/ng.143 URL [本文引用: 1]