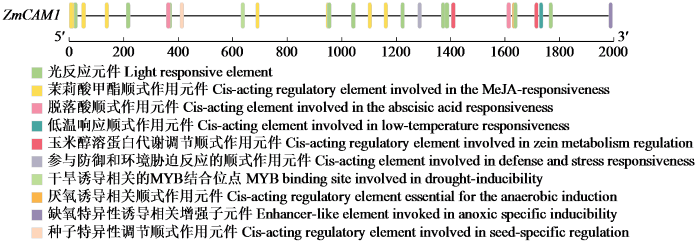钙离子(Ca2+)是生物体内普遍存在的第二信使,在植物的生长发育和应对各种非生物胁迫的反应中发挥着关键作用[1]。钙调蛋白(CaM)是典型的钙离子感受器。典型的CaM蛋白由148个氨基酸组成,是一种酸性蛋白,相对分子量为16.7~16.8 kDa,等电点4.0。它包含4个螺旋―环―螺旋构成的EF-hand结构域[2-3]。CaM在不同真核生物中具有高度的保守性,植物与脊椎动物和酵母的序列同源性分别为91%和61%,植物和藻类之间的序列保守率在84%~100%[4]。在不同植物的基因组测序项目完成后,已在许多植物中鉴定出CaM基因。例如,在拟南芥(Arabidopsis thaliana L.)中发现7种CaM,包含4种蛋白亚型[5]。虽然CaM本身不具备催化活性,但当EF-hand结构域中的EF环与Ca2+结合后,CaM进入活化状态,暴露出富含蛋氨酸的疏水区域[6]。该区域与靶蛋白相互作用,对钙信号进行解码,以响应细胞内Ca2+浓度的短暂变化[7-8]。CaM靶蛋白种类众多且具有广泛的细胞功能,包括信号蛋白(激酶和磷酸酶)、转录因子、细胞骨架相关蛋白、转运蛋白和通道蛋白等。研究[9]发现,CaM在植物生长发育过程以及植物应对环境胁迫的反应中起着重要作用。例如,在玉米种子发芽阶段,CaM基因通过调节线粒体中的琥珀酸脱氢酶活性,调控玉米幼苗对高温的适应能力,并且参与了玉米叶片的抗氧化保护机制[10-11]。在拟南芥花粉萌发及花粉管伸长过程中,功能缺失突变体CaM2的花粉萌发率降低[11]。棉花GhCaM7基因通过调节活性氧(ROS)的产生促进了棉纤维的伸长[12]。在大豆中过表达GmCaM4基因增强了其对3种植物病原菌的抗性,并提高了其对高盐条件的耐受性[13]。
玉米作为一种重要的粮食和饲料作物,容易受到各种非生物胁迫的影响,如涝渍、干旱等。本研究基于玉米自交系B73干旱胁迫转录组表达谱中的差异表达基因,筛选并克隆了一个干旱胁迫后在根系中高水平表达的CaM基因(ZmCaM1),对该蛋白理化性质、保守基序、系统进化树、互作蛋白预测及验证、启动子顺式作用元件、时空表达和干旱胁迫及涝渍胁迫下的表达模式等进行了分析。本文旨在揭示ZmCaM1基因在玉米生长发育以及非生物胁迫中的作用,为进一步探讨该基因的调控机制提供理论支持。
1 材料与方法
1.1 试验材料
TA克隆试剂盒、RNA提取试剂盒、反转录试剂盒以及荧光定量PCR试剂盒均由南京诺唯赞生物有限公司提供;Rosetta2(DE3)感受态、Y2H和Y187酵母感受态由上海昂羽生物技术有限公司提供;谷胱甘肽―琼脂糖凝胶由北京索莱宝科技有限公司提供;所有的引物由生工生物工程(上海)股份有限公司负责合成。
以玉米自交系B73为材料,选择籽粒正常且大小饱满的种子。将种子放入无菌水中于26 ℃浸种12 h,种子吸收足够的水分后,将其播种在塑料杯中进行沙培,每杯放入5粒种子,然后放在组培室,26 ℃培养,待玉米生长至3叶1心时,挑选长势一致的幼苗分别进行涝渍、干旱胁迫处理,将玉米幼苗表面保持1 cm左右的积水进行涝渍处理,利用15%的PEG-6000处理玉米幼苗来模拟干旱胁迫,处理12 h后采集玉米叶和根的组织样品,对照组(CK)为正常条件下生长的植株,每个样本3个重复,冻存于-80 ℃液氮,后续用于RNA提取。另外将玉米种子播种在营养土:泥土=1:2的黑桶中,待玉米长至相应时期,采集玉米根(R)、茎(S)、叶(L)、果穗和雄穗等组织以及出苗期(VE)、3叶期(V3)、7叶期(V7)和吐丝期(R1)等生长时期的样品,-80 ℃保存,每个样本3个重复,后续用于RNA提取。
1.2 玉米RNA的提取
将取好的玉米叶片放入研钵中磨碎,研磨过程中需及时添加液氮以免样品冻融。研磨成粉末的样品放于2 mL离心管中,加入500 µL RNA- easy、200 μL的RNase-free ddH2O混匀。冰上静置10 min。11 000转/min,4 ℃离心10 min。取400 µL上清液放入新的1.5 mL离心管中。加入等体积的异丙醇,上下颠倒,4 ℃静置10 min,11 000转/min离心10 min。加入500 µL 75%乙醇,8000转/min离心3 min(此步骤重复2次)。加入30~50 µL RNase-free ddH2O水溶解,-80 ℃保存。
1.3 ZmCaM1基因克隆
在NCBI查找玉米数据库中ZmCaM1基因(Zm00001d007194)cDNA序列。使用Oligo7设计特异性扩增引物。PCR反应体系:2×Phanta Max Master Mix 25 µL,cDNA 1 µL,上、下游引物各1 µL,ddH2O 22 µL。PCR反应程序为95 ℃,3 min;95 ℃,15 s,60 ℃退火30 s,72 ℃延伸30 s,35个循环;72 ℃延伸5 min。纯化PCR产物,使用5 min TA/Blunt-Zero Cloning Kit试剂盒进行TA克隆,挑取单克隆进行鉴定,将鉴定为阳性的菌液送至生工生物工程(上海)股份有限公司进行测序验证。
1.4 酵母双杂交和蛋白纯化的载体构建
利用诺唯赞GE Design的在线引物设计网站设计引物,将PCR扩增得到的ZmAP1G1、ZmAP1G2、ZmRAD4、ZmCaM1编码序列(CDS)进行测序,通过同源重组反应将测序正确的CDS分别连接到pGBKT7、pGADT7、pGEX-4T-1载体上,鉴定后获得pGBKT7-ZmCaM1、pGADT7-ZmAP1G1、pGADT7-ZmAP1G2、pGADT7-ZmRAD4和pGEX- 4T-1-GST-ZmCaM1。
1.5 生物信息学分析
使用Expasy(
1.6 酵母双杂交试验
将构建好的pGBKT7-ZmCaM1分别与pGADT7-ZmAP1G1、pGADT7-ZmAP1G2、pGADT7-ZmRAD4转入Y2H和Y187酵母感受态中,在SD/-Trp/-Leu培养基上融合培养后,将鉴定为阳性的酵母菌落扩大培养,在OD600达到0.4时用0.9% NaCl溶液进行梯度稀释,取5 µL菌液滴定在SD/-Trp/-Leu/-His/-Ade/3-AT(2.5 mmol/L)培养基上,观察菌落生长情况。
1.7 蛋白原核表达分析及GST标签蛋白的纯化
将构建好的pGEX-4T-1-ZmCaM1转化Rosetta2(DE3),接种单个菌落到含有相应抗生素的5 mL LB培养基中,37 ℃下以200转/min摇动培养过夜,按1:100的比例扩大培养,37 ℃,150转/min震荡培养3~5 h,直到OD600达到0.5~0.8,取1 mL培养物移液至离心管中作为0 h对照,加入IPTG,使各管IPTG最终浓度分别为0.1 mmol/L和0.5 mmol/L,32 ℃与18 ℃振荡培养诱导3~5 h,收集菌体,20 mL PBS缓冲液重悬菌体,并利用超声波进行细胞破碎(开6 s,关6 s,总时间15 min),将表达的蛋白全部释放到溶液中,各取20 μL蛋白溶液分别加入5 μL 5×蛋白电泳上样缓冲液,置沸水浴中煮10 min,最后使用10% SDS聚丙烯酰胺凝胶电泳对蛋白样品进行分析。蛋白纯化步骤参考谷胱甘肽―琼脂糖凝胶(Solarbio公司)说明纯化ZmCaM1蛋白,并进行SDS聚丙烯酰胺凝胶电泳分析。
1.8 ZmCaM1表达分析
使用HiScript 1st Strand cDNA Synthesis Kit试剂盒反转录cDNA,反转录步骤如下。
第1步反应体系:4×gDNA wiper Mix 4 µL,模板RNA 2 µL,RNase-free ddH2O 10 μL,42 ℃,2 min。
第2步反应体系:5×Hi Script Ⅱ qRT Super MixⅡ 4 μL,第1步的反应液16 μL,50 ℃,15 min;85 ℃,15 s。利用Oligo7设计定量引物,以cDNA为模板,利用诺唯赞的SYBR Green进行实时荧光定量PCR分析,qRT-PCR扩增程序:95 ℃预变性30 s,随后进行35个循环:95 ℃变性5 s,60 ℃退火34 s。以ZmActin1为内参基因,利用2-ΔΔCt法计算出基因的相对表达量。所用引物见表1。
表1 PCR和qRT-PCR引物的信息
Table 1
| 引物名称Primer name | 基因ID Gene ID | 上游引物Forward primer (5′-3′) | 下游引物Reverse primer (5′-3′) |
|---|---|---|---|
| ZmCaM1 | Zm00001d007194 | ATGGCGGACCAGCTCACC | TCACTTGGCCATCATGACCTT |
| qZmCaM1 | Zm00001d007194 | TCAAATCGCTCCCTGCCTCT | GACCCAGTGATCGCATGACA |
| BK-CaM1 | Zm00001d007194 | AGGCCGAATTCCCGGGGATCCAT GGCGGACCAACTCACCG | CCGCTGCAGGTCGACGGATCCTCAC TTGGCCATCATGACCTT |
| AD-AP1G1 | Zm00001d045195 | GCCATGGAGGCCAGTGAATTCATGG ACCTCGCCATCAATCC | ACGATTCATCTGCAGCTCGAGCTACA ACCCAGCAGGAAAGTTGC |
| AD-AP1G2 | Zm00001d036305 | GCCATGGAGGCCAGTGAATTCAT GGACCTCGCCATCAACC | ACGATTCATCTGCAGCTCGAGCTACAAC CCAGAAGGAAAGTTGC |
| AD-RAD4 | Zm00001d032245 | GCCATGGAGGCCAGTGAATTCA TGCGGCGGACGAGGAGC | ACGATTCATCTGCAGCTCGAGTTACAAC TCCTCTACTTGGATAGAAAAAC |
| GST-CaM1 | Zm00001d007194 | GATCTGGTTCCGCGTGGATCCATGGCGG ACCAGCTCACC | GTCACGATGCGGCCGCTCGAGTCA CTTGGCCATCATGACCTT |
| qZmActin | Actin | TTGCTATCCAGGCTGTTCTT | CGACCTCAGCAGCGCGGTCA |
2 结果与分析
2.1 ZmCaM1基因克隆
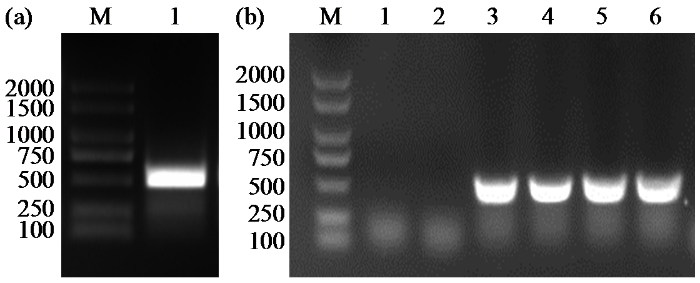
图1
图1
ZmCaM1基因的CDS扩增(a)和菌落PCR扩增(b)
1~6代表6个不同的单克隆菌落;M:2000 bp DNA分子量标记。
Fig.1
CDS amplification of ZmCaM1 gene (a) and the colony PCR amplification (b)
Lines 1-6, six different colonies; M: 2000 bp DNA marker.
2.2 ZmCaM1蛋白理化性质分析
对ZmCaM1蛋白理化性质进行分析,结果(表2)显示,ZmCaM1蛋白氨基酸数目为149,分子式为C722H1135N189O253S10,分子量为16 831.67 kDa,带正电荷氨基酸残基总数(Arg+Lys)为15个,带负电荷氨基酸残基总数(Asp+Glu)为38个。脂肪指数为70.07,平均疏水系数为-0.602,半衰期为30.00 h,理论等电点为4.11,属于酸性蛋白质。不稳定系数23.23,推测ZmCaM1蛋白为不稳定蛋白。
表2 ZmCaM1蛋白理化性质
Table 2
| 一级结构特征 Characteristics of primary structure | 预测结果 Prediction result |
|---|---|
| 氨基酸数量Number of amino acids | 149 |
| 分子量Molecular weight (kDa) | 16 831.67 |
| 分子式Molecular formula | C722H1135N189O253S10 |
| 带正电荷残基总数 Total number of positively charged residues | 15 |
| 带负电荷残基总数 Total number of negatively charged residues | 38 |
| 平均疏水性Average hydrophobicity | -0.602 |
| 脂肪指数Aliphatic index | 70.07 |
| 不稳定系数Instability coefficient | 23.23 |
| 半衰期Estimated half-life (h) | 30.00 |
| 等电点Isoelectric point | 4.11 |
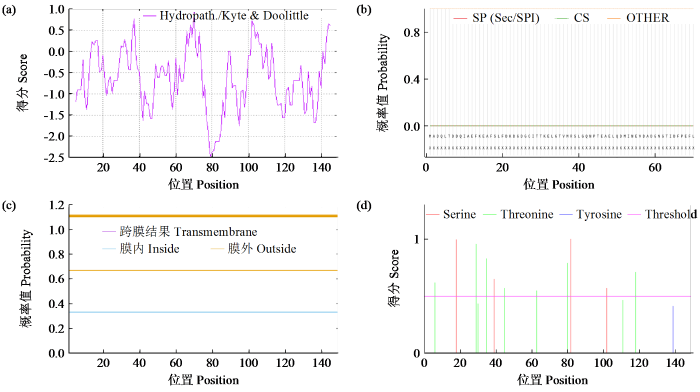
2.3 ZmCaM1蛋白信号肽、亲疏水性和磷酸化位点分析
图2
图2
ZmCaM1蛋白亲水性与疏水性预测(a)、蛋白信号肽预测(b)、蛋白跨膜结构预测(c)及蛋白磷酸化位点预测(d)
Fig.2
ZmCaM1 protein hydrophilicity/hydrophobicity prediction (a), signal peptide prediction (b), transmembrane structure prediction (c) and phosphorylation sites prediction (d)
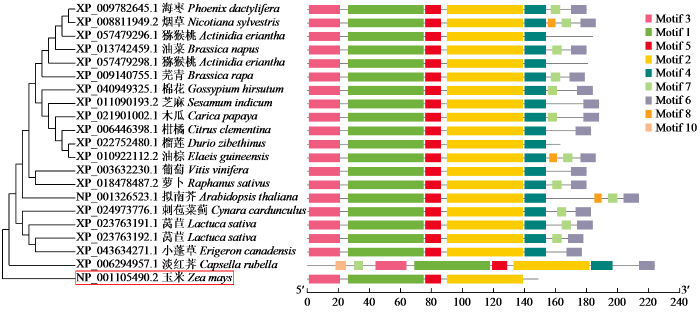
2.4 ZmCaM1进化树构建和保守基序分析
为了研究ZmCaM1蛋白的进化关系,我们选取了20个ZmCaM1同源蛋白进行分析。序列比对结果(图3)显示,ZmCaM1蛋白序列具有高度的保守性,有较长的保守区域和高氨基酸相似度。进化分析(图4)显示,ZmCaM1与淡红荠XP_006294957.1关系最近,其次是小蓬草XP_043634271.1,而与海枣XP_009782645.1亲缘关系最远。10个共有基序命名为Motif 1~Motif 10(表3),其中Motif 1、Motif 2、Motif 3和Motif 5在大多数同源蛋白中存在。前149个氨基酸上的基序排列和其他20种同源蛋白中非常相似。以上结果显示ZmCaM1蛋白在其序列特征上与其他物种中已鉴定的CaM高度相似,推测ZmCaM1蛋白的这些保守序列对于其生物学功能的正常发挥至关重要。
图3
图3
ZmCaM1氨基酸序列与其他20个同源序列比对
蓝色:相似性100%;粉色:相似性≥75%;黄色:相似性≥50%。
Fig.3
Comparison of ZmCaM1 amino acid sequences with other 20 homologous sequences
Blue: Similarity 100%; Pink: Similarity greater than or equal to 75%; Yellow: Similarity greater than or equal to 50%.
图4
图4
ZmCaM1进化树构建和保守结构域
Fig.4
Construction of evolutionary tree and conserved domains of ZmCaM1
表3 ZmCaM蛋白保守基序
Table 3
| Motif编号Motif number | Motif序列Motif sequence |
|---|---|
| Motif 1 | GCITTKELGTVMRSLGQNPTEAELQDMINEVDADGNGTIDFPEFLNLMAR |
| Motif 2 | FRVFDKDQNGFISAAELRHVMTNLGEKLTDEEVDEMIREADVDGDGQINY |
| Motif 3 | MADQLTDDQISEFKEAFSLFD |
| Motif 4 | EEFVKVMMAKRRRKR |
| Motif 5 | KMKDTDSEEEL |
| Motif 6 | PSSSTERKEERRRRKSHCRIL |
| Motif 7 | HNPSQP |
| Motif 8 | GIWKYAASKLLEIRKSHKRFP |
| Motif 9 | IQEKRRGSEPERSPSPQK |
| Motif 10 | LNHTNV |
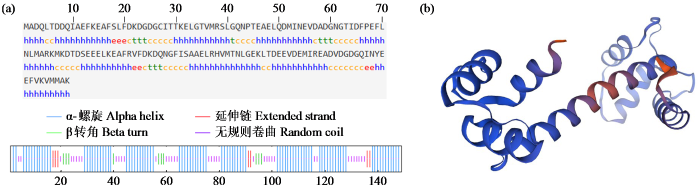
2.5 ZmCaM1蛋白结构分析
图5
图5
ZmCaM1蛋白二级结构(a)与三级结构(b)
Fig.5
Secondary structure (a) and tertiary structure (b) of ZmCaM1 protein
2.6 ZmCaM1基因启动子序列分析
对ZmCaM1基因启动子区域包含的顺式作用元件进行分析,结果(图6)显示,在ZmCaM1基因转录起始位点的上游2000 bp内存在多种顺式作用元件。其中包括有关干旱响应的MYB结合位点和低温响应元件LTR等,说明ZmCaM1基因可能参与干旱和低温等非生物胁迫的响应。其次,还发现有茉莉酸甲酯响应元件TGACG-motif和脱落酸响应元件ABRE,说明ZmCaM1基因可能受激素的调控。再次,还发现了光响应元件GT1-motif,提示ZmCaM1基因可能响应光的调控。综上所述,ZmCaM1基因可能在激素反应、非生物胁迫应答以及光信号处理等多种途径中起到关键作用。
图6
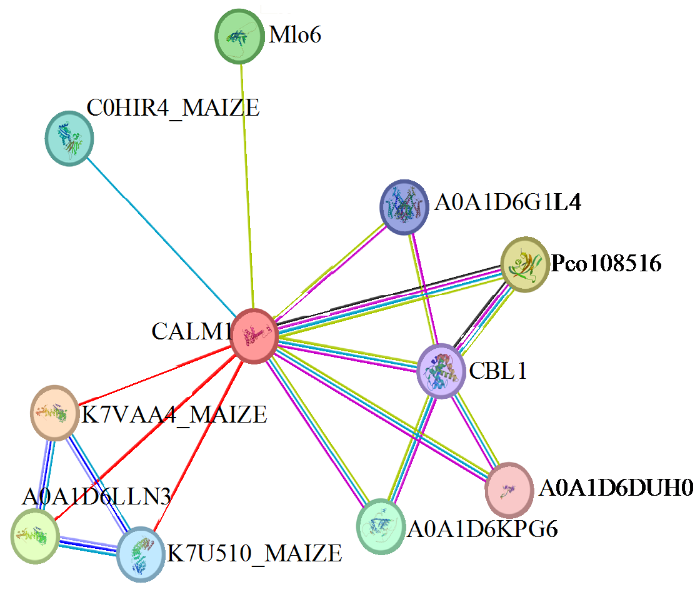
2.7 ZmCaM1蛋白互作分析
STRING互作蛋白预测结果(图7)显示,ZmCaM1共与10个蛋白互作,包括3个AP-1复合亚基γ(K7VAA4_MAIZE、A0A1D6LLN3、K7U510_MAIZE)、肽基―脯氨酰顺式/反式异构酶(Pco108516)、类MLO蛋白(Mlo6)、胚胎缺陷14蛋白(C0HIR4_MAIZE)、钙调磷酸酶B类似蛋白1(CBL1)、双孔钙通道蛋白1(A0A1D6G1L4)、驱动蛋白KCBP(A0A1D6DUH0)、DNA修复蛋白RAD4(A0A1D6KPG6)。
图7
2.8 ZmCaM1互作蛋白的验证
为了进一步验证ZmCaM1互作蛋白的预测结果,将克隆得到的ZmCaM1的CDS序列连接到pGBKT7(BD)载体上,并将ZmAP1G1、ZmAP1G2和ZmRAD4的CDS序列分别连接到pGADT7(AD)载体上,进行酵母双杂交点对点试验。试验结果(图8)显示,试验组pGBKT7-ZmCaM1/pGADT7- ZmAP1G1、pGBKT7-ZmCaM1/pGADT7-ZmAP1G2、pGBKT7-ZmCaM1/pGADT7-ZmRAD4和对照组pGBKT7-p53/pGADT7-T、pGBKT7-Lam/pGADT7-T、pGBKT7-ZmCaM1/pGADT7在SD/-Trp/-Leu培养基上均能正常生长。而在SD/-Trp/-Leu/-His/-Ade/ 3-AT(2.5 mmol/L)培养基上,试验组pGBKT7- ZmCaM1/pGADT7-ZmRAD4和阳性对照组pGBKT7-p53/pGADT7-T能正常生长,试验组pGBKT7-ZmCaM1/pGADT7-ZmAP1G1、pGBKT7- ZmCaM1/pGADT7-ZmAP1G2和阴性对照组pGBKT7-Lam/pGADT7-T、pGBKT7-ZmCaM1/ pGADT7均不能正常生长。上述结果表明,ZmCaM1与ZmRAD4之间存在相互作用,而与ZmAP1G1、ZmAP1G2之间不存在相互作用。
图8
图8
酵母双杂交验证ZmCaM1互作蛋白
Fig. 8
The interaction validation between ZmCaM1 and other proteins by yeast two-hybrid
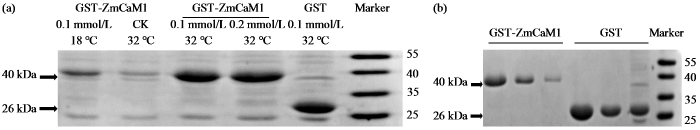
2.9 GST-ZmCaM1蛋白原核表达分析及纯化
为了优化GST-ZmCaM1蛋白诱导条件,在不同温度和IPTG浓度下进行了GST-ZmCaM1蛋白的诱导试验。结果(图9a)显示,GST-ZmCaM1蛋白在32 ℃条件下的表达效果较18 ℃更佳,条带更加清晰。GST-ZmCaM1蛋白在0.1 mmol/L IPTG条件下的表达效果较0.2 mmol/L更佳。综上所述,GST- ZmCaM1蛋白在32 ℃和0.1 mmol/L IPTG条件下的诱导表达效果最佳。随后,我们在上述条件下使用谷胱甘肽―琼脂糖树脂纯化了GST-ZmCaM1蛋白和对照GST蛋白。SDS-PAGE分析结果(图9b)显示,GST-ZmCaM1蛋白大约在40 kDa处且为单一条带,表明GST-ZmCaM1蛋白已成功纯化。
图9
图9
GST-ZmCaM1蛋白原核表达分析(a)及蛋白纯化结果(b)
Fig.9
Analysis of prokaryotic expression (a) and purification (b) of ZmCaM1
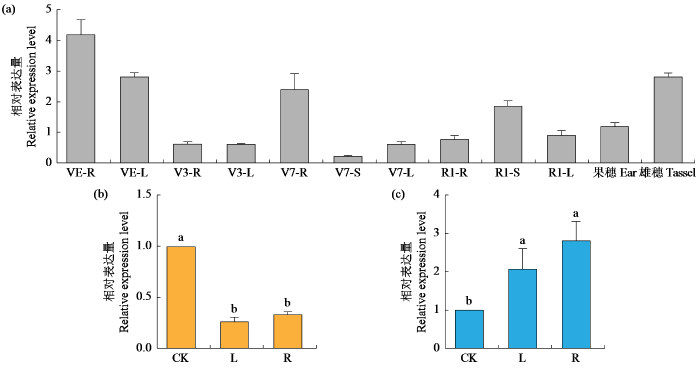
2.10 玉米ZmCaM1基因表达模式分析
通过qRT-PCR技术分析ZmCaM1基因在玉米不同组织和生长阶段的表达量,结果(图10a)显示,在玉米自交系B73中,在根、茎、叶、雌穗和雄穗等组织以及VE、V3、V7和R1等时期中均有表达。其中,在VE期的根和叶中表达量明显高于其他组织。这一结果表明该基因可能在玉米种子萌发这一发育过程中发挥重要功能。
图10
图10
ZmCaM1基因在不同时期组织(a)中及涝渍(b)与干旱胁迫处理下(c)的相对表达量
不同小写字母表示P < 0.05水平差异显著。
Fig.10
The relative expression levels of ZmCaM1 gene in tissues at various stages (a), and under waterlogging (b) and drought stress treatments (c)
The different lowercase letters indicate significant difference at P < 0.05 level.
对干旱胁迫和涝渍胁迫后ZmCaM1基因在玉米根和叶中的表达量进行分析,结果(图10b~c)表明,干旱处理后,ZmCaM1基因在叶和根中表达水平显著上升;涝渍处理后,ZmCaM1基因在叶和根中表达水平显著下降。推测ZmCaM1基因可能在玉米遭受干旱和涝渍胁迫时,参与了相关的信号转导途径。
3 讨论
在众多植物物种中,CaM基因家族的成员已被鉴定,其编码的蛋白质结构高度保守[15]。本研究从玉米自交系B73中克隆了ZmCaM1基因。蛋白结构分析显示,ZmCaM1蛋白具有4个EF-hand结构域,与拟南芥、水稻、大麦中的CaM结构相似。系统发育分析显示,ZmCaM1与十字花科芥属植物淡红荠的CaM5蛋白关系最近。这些发现证实ZmCaM1蛋白具有钙调蛋白的典型保守特征。在细胞内信号转导机制中,磷酸化修饰是蛋白质活性和功能的基本调节方式,具有至关重要的作用。ZmCaM1蛋白含有丝氨酸和苏氨酸磷酸化位点,这表明ZmCaM1可能通过参与信号转导过程,在不同生物学进程和分子功能上起到重要的调节作用。通过原核表达诱导条件的优化,GST-ZmCaM1蛋白的最佳诱导条件为32 ℃和0.1 mmol/L IPTG,最终成功纯化了GST-ZmCaM1蛋白。这一优化和纯化过程为后续的蛋白活性分析及Pull-Down互作验证等试验奠定了坚实的基础。
Ca2+是植物生长发育和非生物胁迫应答中的关键第二信使[16]。CaM作为一种广泛研究的钙感受器,在植物的生长发育中起着重要作用[17]。例如,AtCML19/AtCML20与AtTON1的互作调节植物维管组织的形成[18]。此外,CaM也在植物激素响应过程中起作用。如在玉米中,ZmSAUR1与CaM互作调控生长素介导的细胞伸长[7]。在本研究中,通过qRT-PCR方法测定了ZmCaM1基因在不同时期和不同组织的表达量,结果显示该基因在VE期根和叶中表达量明显高于其他组织。揭示其在玉米种子萌发这一生长发育过程中发挥重要作用,间接证实CaM基因在不同物种中的保守性及其在玉米生长发育过程中的重要作用。ZmCaM1基因启动子区顺式作用元件分析显示,ZmCaM1基因启动子中包含多个激素和逆境响应元件,如MeJA响应元件TGACG-motif、ABA响应元件ABRE和干旱响应的MYB结合位点MBS等。在ZmCaM1互作蛋白预测和酵母双杂交试验中发现,ZmCaM1蛋白与DNA修复蛋白RAD4存在相互作用,而DNA修复蛋白RAD4则与DNA修复途径密切相关。基于上述分析结果,可以推测在玉米中,激素信号如甲基茉莉酸(MeJA)和脱落酸(ABA)可能通过调节ZmCaM1基因的表达来影响ZmCaM1蛋白与DNA修复蛋白RAD4的相互作用。这种调控可能进一步作用于植物细胞的DNA修复途径,以维持细胞稳定性,并减轻或消除环境胁迫对玉米的不利影响。
众多研究[19-
综上,本研究通过ZmCaM1克隆、功能和结构的分析、原核表达等研究,揭示了ZmCaM1与多种非生物胁迫以及生长发育过程相关。通过筛选和验证ZmCaM1的互作蛋白及其功能分析,初步提出了ZmCaM1可能提高玉米抗逆性的分子机制,为进一步完善玉米钙信号转导网络提供了新的理论依据。对ZmCaM1基因功能和分子机制研究将推动钙信号途径在提高玉米抗逆性方面的研究和应用,为改良玉米抗逆品种提供了新的理论基础。
4 结论
ZmCaM1氨基酸数目为149个,蛋白大小约为17 kDa,其结构域和保守基序与其他植物中的CaM高度相似。qRT-PCR分析发现,在VE期ZmCaM1在根和叶中的表达水平明显高于其他组织,在干旱胁迫条件下,ZmCaM1基因表达水平上调,而在涝渍胁迫后下调,这表明ZmCaM1可能在玉米的生长发育和逆境应答中发挥重要作用。通过酵母双杂交筛选,鉴定出ZmCaM1与DNA修复蛋白RAD4发生互作。
参考文献
The evolution of Ca2+ signalling in photosynthetic eukaryotes
Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin
Identification,characterization, and expression analysis of calmodulin and calmodulin-like genes in grapevine (Vitis vinifera) reveal likely roles in stress responses
DOI:S0981-9428(18)30254-7
PMID:29908490
[本文引用: 1]

Calcium (Ca) is an ubiquitous key second messenger in plants, where it modulates many developmental and adaptive processes in response to various stimuli. Several proteins containing Ca binding domain have been identified in plants, including calmodulin (CaM) and calmodulin-like (CML) proteins, which play critical roles in translating Ca signals into proper cellular responses. In this work, a genome-wide analysis conducted in Vitis vinifera identified three CaM- and 62 CML-encoding genes. We assigned gene family nomenclature, analyzed gene structure, chromosomal location and gene duplication, as well as protein motif organization. The phylogenetic clustering revealed a total of eight subgroups, including one unique clade of VviCaMs distinct from VviCMLs. VviCaMs were found to contain four EF-hand motifs whereas VviCML proteins have one to five. Most of grapevine CML genes were intronless, while VviCaMs were intron rich. All the genes were well spread among the 19 grapevine chromosomes and displayed a high level of duplication. The expression profiling of VviCaM/VviCML genes revealed a broad expression pattern across all grape organs and tissues at various developmental stages, and a significant modulation in biotic stress-related responses. Our results highlight the complexity of CaM/CML protein family also in grapevine, supporting the versatile role of its different members in modulating cellular responses to various stimuli, in particular to biotic stresses. This work lays the foundation for further functional and structural studies on specific grapevine CaMs/CMLs in order to better understand the role of Ca-binding proteins in grapevine and to explore their potential for further biotechnological applications.Copyright © 2018 Elsevier Masson SAS. All rights reserved.
Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action
DOI:10.1074/jbc.275.5.3137
PMID:10652297
[本文引用: 1]

The use of (35)S-labeled calmodulin (CaM) to screen a corn root cDNA expression library has led to the isolation of a CaM-binding protein, encoded by a cDNA with sequence similarity to small auxin up RNAs (SAURs), a class of early auxin-responsive genes. The cDNA designated as ZmSAUR1 (Zea mays SAURs) was expressed in Escherichia coli, and the recombinant protein was purified by CaM affinity chromatography. The CaM binding assay revealed that the recombinant protein binds to CaM in a calcium-dependent manner. Deletion analysis revealed that the CaM binding site was located at the NH(2)-terminal domain. A synthetic peptide of amino acids 20-45, corresponding to the potential CaM binding region, was used for calcium-dependent mobility shift assays. The synthetic peptide formed a stable complex with CaM only in the presence of calcium. The CaM affinity assay indicated that ZmSAUR1 binds to CaM with high affinity (K(d) approximately 15 nM) in a calcium-dependent manner. Comparison of the NH(2)-terminal portions of all of the characterized SAURs revealed that they all contain a stretch of the basic alpha-amphiphilic helix similar to the CaM binding region of ZmSAUR1. CaM binds to the two synthetic peptides from the NH(2)-terminal regions of Arabidopsis SAUR-AC1 and soybean 10A5, suggesting that this is a general phenomenon for all SAURs. Northern analysis was carried out using the total RNA isolated from auxin-treated corn coleoptile segments. ZmSAUR1 gene expression began within 10 min, increased rapidly between 10 and 60 min, and peaked around 60 min after 10 microM alpha-naphthaleneacetic acid treatment. These results indicate that ZmSAUR1 is an early auxin-responsive gene. The CaM antagonist N-(6-aminohexyl)5-chloro-1-naphthalenesulfonamide hydrochloride inhibited the auxin-induced cell elongation but not the auxin-induced expression of ZmSAUR1. This suggests that calcium/CaM do not regulate ZmSAUR1 at the transcriptional level. CaM binding to ZmSAUR1 in a calcium-dependent manner suggests that calcium/CaM regulate ZmSAUR1 at the post-translational level. Our data provide the first direct evidence for the involvement of calcium/CaM-mediated signaling in auxin-mediated signal transduction.
Nitric oxide functions as a signal and acts upstream of AtCaM3 in thermotolerance in Arabidopsis seedlings
DOI:10.1104/pp.110.160424
PMID:20576787
[本文引用: 2]

To characterize the role of nitric oxide (NO) in the tolerance of Arabidopsis (Arabidopsis thaliana) to heat shock (HS), we investigated the effects of heat on three types of Arabidopsis seedlings: wild type, noa1(rif1) (for nitric oxide associated1/resistant to inhibition by fosmidomycin1) and nia1nia2 (for nitrate reductase [NR]-defective double mutant), which both exhibit reduced endogenous NO levels, and a rescued line of noa1(rif1). After HS treatment, the survival ratios of the mutant seedlings were lower than those of wild type; however, they were partially restored in the rescued line. Treatment of the seedlings with sodium nitroprusside or S-nitroso-N-acetylpenicillamine revealed that internal NO affects heat sensitivity in a concentration-dependent manner. Calmodulin 3 (CaM3) is a key component of HS signaling in Arabidopsis. Real-time reverse transcription-polymerase chain reaction analysis after HS treatment revealed that the AtCaM3 mRNA level was regulated by the internal NO level. Sodium nitroprusside enhanced the survival of the wild-type and noa1(rif1) seedlings; however, no obvious effects were observed for cam3 single or cam3noa1(rif1) double mutant seedlings, suggesting that AtCaM3 is involved in NO signal transduction as a downstream factor. This point was verified by phenotypic analysis and thermotolerance testing using seedlings of three AtCaM3-overexpressing transgenic lines in an noa1(rif1) background. Electrophoretic mobility-shift and western-blot analyses demonstrated that after HS treatment, NO stimulated the DNA-binding activity of HS transcription factors and the accumulation of heat shock protein 18.2 (HSP18.2) through AtCaM3. These data indicate that NO functions in signaling and acts upstream of AtCaM3 in thermotolerance, which is dependent on increased HS transcription factor DNA-binding activity and HSP accumulation.
Plant calmodulins and calmodulin- related proteins: multifaceted relays to decode calcium signals
The CBL-CIPK Ca2+-decoding signaling network: function and perspectives
Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins
Calmodulin-related CML24 interacts with ATG4b and affects autophagy progression in Arabidopsis
Genomics and evolutionary aspect of calcium signaling event in calmodulin and calmodulin- like proteins in plants
Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis
Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis
Characterization of GmCaMK1, a member of a soybean calmodulin-binding receptor- like kinase family
DOI:10.1016/j.febslet.2010.10.059
PMID:21056039
[本文引用: 1]

Calmodulin(CaM)-regulated protein phosphorylation forms an important component of Ca(2+) signaling in animals but is less understood in plants. We have identified a CaM-binding receptor-like kinase from soybean nodules, GmCaMK1, a homolog of Arabidopsis CRLK1. We delineated the CaM-binding domain (CaMBD) of GmCaMK1 to a 24-residue region near the C-terminus, which overlaps with the kinase domain. We have demonstrated that GmCaMK1 binds CaM with high affinity in a Ca(2+)-dependent manner. We showed that GmCaMK1 is expressed broadly across tissues and is enriched in roots and developing nodules. Finally, we examined the CaMBDs of the five-member GmCaMK family in soybean, and orthologs present across taxa.Copyright © 2010 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner
Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense
DOI:10.1111/j.1365-313X.2005.02435.x
PMID:15960618
[本文引用: 1]

Glucosinolates are a class of secondary metabolites with important roles in plant defense and human nutrition. To uncover regulatory mechanisms of glucosinolate production, we screened Arabidopsis thaliana T-DNA activation-tagged lines and identified a high-glucosinolate mutant caused by overexpression of IQD1 (At3g09710). A series of gain- and loss-of-function IQD1 alleles in different accessions correlates with increased and decreased glucosinolate levels, respectively. IQD1 encodes a novel protein that contains putative nuclear localization signals and several motifs known to mediate calmodulin binding, which are arranged in a plant-specific segment of 67 amino acids, called the IQ67 domain. We demonstrate that an IQD1-GFP fusion protein is targeted to the cell nucleus and that recombinant IQD1 binds to calmodulin in a Ca(2+)-dependent fashion. Analysis of steady-state messenger RNA levels of glucosinolate pathway genes indicates that IQD1 affects expression of multiple genes with roles in glucosinolate metabolism. Histochemical analysis of tissue-specific IQD1::GUS expression reveals IQD1 promoter activity mainly in vascular tissues of all organs, consistent with the expression patterns of several glucosinolate-related genes. Interestingly, overexpression of IQD1 reduces insect herbivory, which we demonstrated in dual-choice assays with the generalist phloem-feeding green peach aphid (Myzus persicae), and in weight-gain assays with the cabbage looper (Trichoplusia ni), a generalist-chewing lepidopteran. As IQD1 is induced by mechanical stimuli, we propose IQD1 to be novel nuclear factor that integrates intracellular Ca(2+) signals to fine-tune glucosinolate accumulation in response to biotic challenge.
Nitric oxide, stomatal closure, and abiotic stress
Handling calcium signaling: Arabidopsis CaMs and CMLs
Calmodulins and related potential calcium sensors of Arabidopsis