作物杂志,2023, 第3期: 246–253 doi: 10.16035/j.issn.1001-7283.2023.03.034
稻瘟病菌类SRRM1转录因子的功能分析
- 黑龙江八一农垦大学,163319,黑龙江大庆
Functional Analysis of SRRM1-Like Transcription Factor of Magnaporthe grisea
Qiu Kaihua( ), Fang Shumei, Liang Xilong(
), Fang Shumei, Liang Xilong( )
)
- Heilongjiang Bayi Agricultural University, Daqing 163319, Heilongjiang, China
摘要:
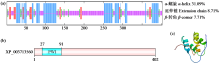
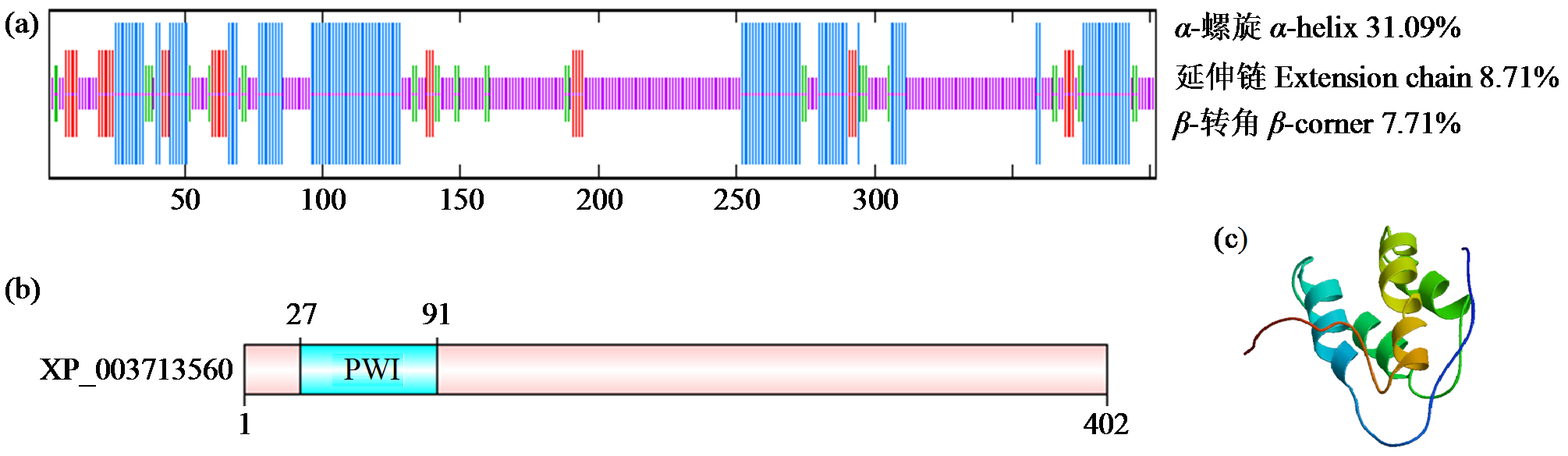
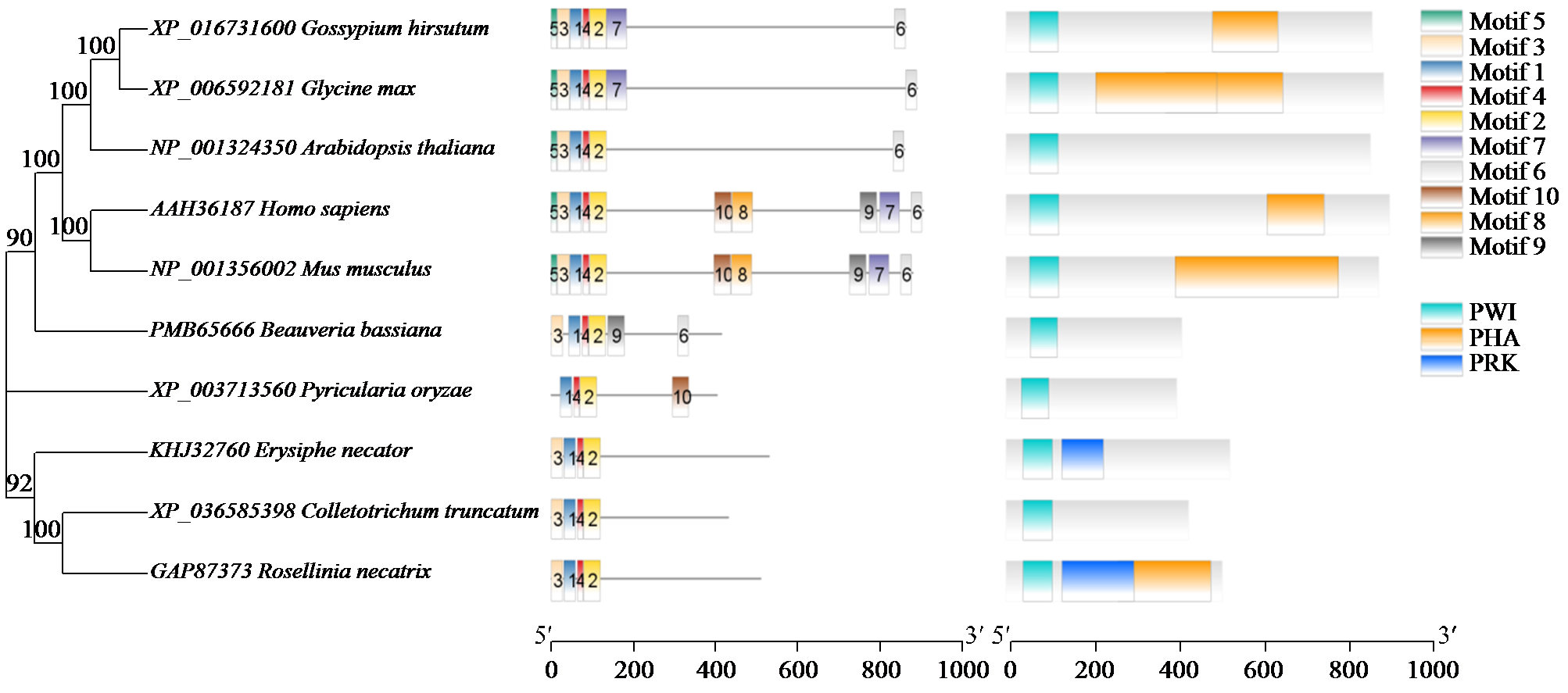
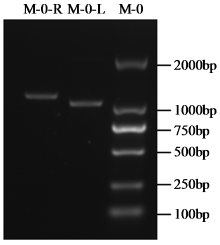
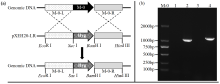
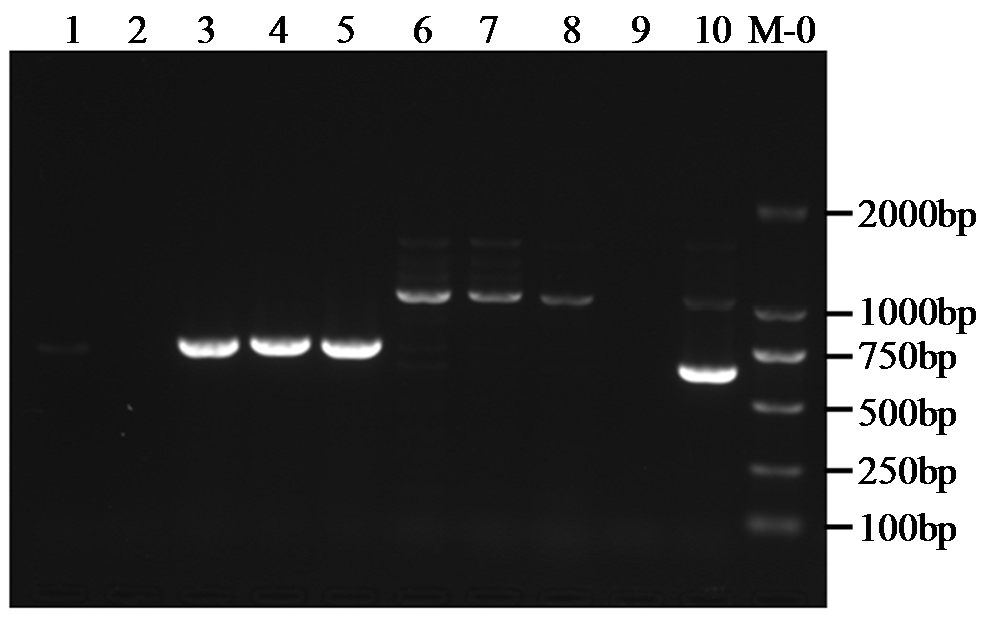
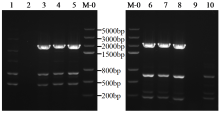
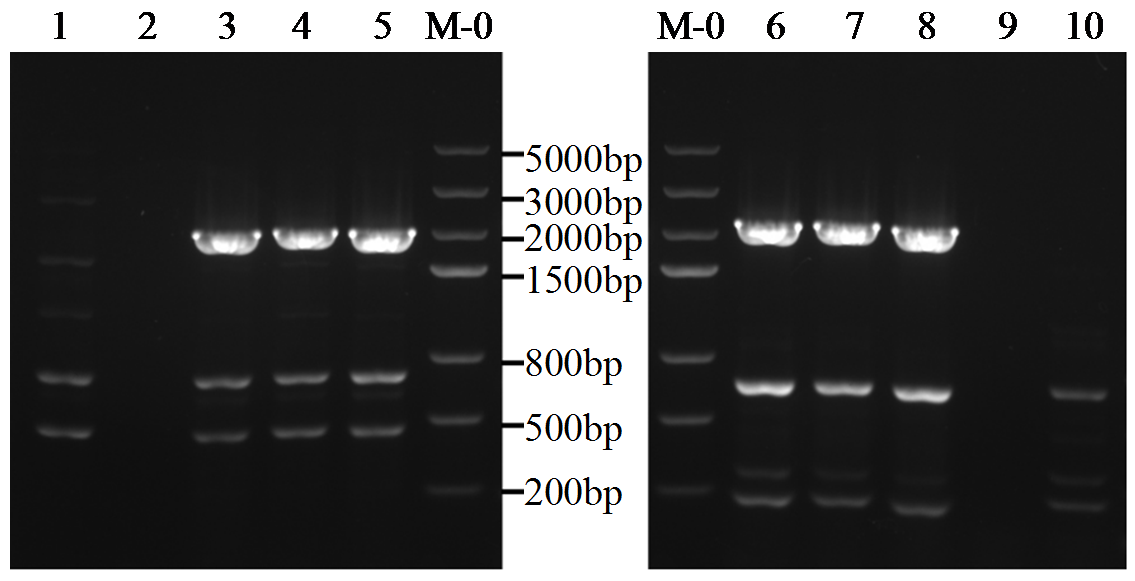
转录因子在稻瘟病菌(Magnaporthe grisea)的生长发育、逆境适应以及致病性中发挥着重要作用。为明确类SRRM1转录因子调控的pre-mRNA剪接与稻瘟病菌生长发育和逆境适应的关系,采用以基因重组原理为基础的基因定点敲除技术获取类SRRM1基因缺失菌株,并分析营养生长及逆境适应。结果表明,本研究成功构建敲除载体并敲除了稻瘟病菌Y34菌株中的类SRRM1基因,获得基因缺失突变体。稻瘟病菌中类SRRM1基因缺失未明显影响其营养生长,但对刚果红、十二烷基硫酸钠、山梨醇、NaCl及杀菌剂吡唑醚菌酯逆境胁迫敏感,表现为抗逆性减弱。类SRRM1在稻瘟病菌逆境适应过程中发挥重要作用。
| [1] | Will C L, Lührmann R. Spliceosome structure and function. Cold Spring Harbor Perspectives in Biology, 2011, 3(7):a003707. |
| [2] |
Kelly S M, Corbett A H. Messenger RNA export from the nucleus:a series of molecular wardrobe changes. Traffic, 2009, 10(9):1199-1208.
doi: 10.1111/j.1600-0854.2009.00944.x pmid: 19552647 |
| [3] |
Nino C A, Herissant L, Babour A, et al. mRNA nuclear export in yeast. Chemical Reviews, 2013, 113(11):8523-8545.
doi: 10.1021/cr400002g pmid: 23731471 |
| [4] |
Katahira J. Nuclear export of messenger RNA. Genes, 2015, 6(2):163-184.
doi: 10.3390/genes6020163 pmid: 25836925 |
| [5] | Burge C B, Tuschl T, Sharp P A. Splicing of precursors to mRNAs by the spliceosomes. Cold Spring Harbor Monograph Series, 1999, 37:525-560. |
| [6] |
Jurica M S, Moore M J. Pre-mRNA splicing:awash in a sea of proteins. Molecular Cell, 2003, 12(1):5-14.
doi: 10.1016/S1097-2765(03)00270-3 |
| [7] |
Kastner B, Will C L, Stark H, et al. Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harbor Perspectives in Biology, 2019, 11(11):a032417.
doi: 10.1101/cshperspect.a032417 |
| [8] | Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNAs by the spliceosome. Cold Spring Harbor Monograph Series, 1993, 24:303. |
| [9] |
Cáceres J F, Kornblihtt A R. Alternative splicing:multiple control mechanisms and involvement in human disease. Trends in Genetics, 2002, 18(4):186-193.
doi: 10.1016/s0168-9525(01)02626-9 pmid: 11932019 |
| [10] |
Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA, 1995, 1(7):663-680.
pmid: 7585252 |
| [11] |
Graveley B R. Sorting out the complexity of SR protein functions. RNA, 2000, 6(9):1197-1211.
pmid: 10999598 |
| [12] |
Blencowe B J, Bowman J A L, McCracken S, et al. SR-related proteins and the processing of messenger RNA precursors. Biochemistry and Cell Biology, 1999, 77(4):277-291.
pmid: 10546891 |
| [13] | Wagner S, Chiosea S, Nickerson J A. The spatial targeting and nuclear matrix binding domains of SRm160. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(6):3269-3274. |
| [14] |
Szymczyna B R, Bowman J, McCracken S, et al. Structure and function of the PWI motif:a novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes and Development, 2003, 17(4):461-475.
pmid: 12600940 |
| [15] |
Blencowe B J, Ouzounis C A. The PWI motif:a new protein domain in splicing factors. Trends in Biochemical Sciences, 1999, 24(5):179-180.
pmid: 10322432 |
| [16] |
Wagner S, Chiosea S, Ivshina M, et al. In vitro FRAP reveals the ATP-dependent nuclear mobilization of the exon junction complex protein SRm160. The Journal of Cell Biology, 2004, 164(6):843-850.
doi: 10.1083/jcb.200307002 |
| [17] |
Kataoka N, Yong J, Kim V N, et al. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Molecular Cell, 2000, 6(3):673-682.
pmid: 11030346 |
| [18] |
Kim V N, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science, 2001, 293(5536):1832-1836.
pmid: 11546873 |
| [19] |
Le Hir H, Moore M J, Maquat L E. Pre-mRNA splicing alters mRNP composition:evidence for stable association of proteins at exon-exon junctions. Genes and Development, 2000, 14(9):1098-1108.
pmid: 10809668 |
| [20] |
Le Hir H, Izaurralde E, Maquat L E, et al. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. The European Molecular Biology Organization Journal, 2000, 19(24):6860-6869.
doi: 10.1093/emboj/19.24.6860 |
| [21] |
Lejeune F, Ishigaki Y, Li X, et al. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells:dynamics of mRNP remodeling. The European Molecular Biology Organization Journal, 2002, 21(13):3536- 3545.
doi: 10.1093/emboj/cdf345 |
| [22] |
Beckwith E J, Hernando C E, Polcowñuk S, et al. Rhythmic behavior is controlled by the SRm 160 splicing factor in Drosophila melanogaster. Genetics, 2017, 207(2):593-607.
doi: 10.1534/genetics.117.300139 pmid: 28801530 |
| [23] | McCracken S, Longman D, Johnstone I L, et al. An evolutionarily conserved role for SRm 160 in 3′-end processing that functions independently of exon junction complex formation. Biological Chemistry, 2003, 278(45):44153-44160. |
| [24] |
Long J C, Caceres J F. The SR protein family of splicing factors:master regulators of gene expression. Biochemical Journal, 2009, 417(1):15-27.
doi: 10.1042/BJ20081501 |
| [25] | Jeong S. SR proteins:binders,regulators, and connectors of RNA. Molecules and Cells, 2017, 40(1):1. |
| [26] |
Blencowe B J, Issner R, Nickerson J A, et al. A coactivator of pre-mRNA splicing. Genes and Development, 1998, 12(7):996-1009.
pmid: 9531537 |
| [27] | McCracken S, Longman D, Marcon E, et al. Proteomic analysis of SRm160-containing complexes reveals a conserved association with cohesin. Biological Chemistry, 2005, 280(51):42227-42236. |
| [28] |
Cheng C, Sharp P A. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Molecular Cell Biology, 2006, 26(1):362-370.
doi: 10.1128/MCB.26.1.362-370.2006 |
| [1] | 孟亚轩, 姚旭航, 孙颖琦, 赵心月, 王凤霞, 瓮巧云, 刘颖慧. 主要禾谷类作物DGAT基因家族比较分析[J]. 作物杂志, 2023, (1): 20–29 |
| [2] | 尹桂芳, 段迎, 杨晓琳, 蔡苏云, 王艳青, 卢文洁, 孙道旺, 贺润丽, 王莉花. 苦荞FtC4H基因克隆与生物信息学分析[J]. 作物杂志, 2022, (1): 77–83 |
| [3] | 孟峰, 张亚玲, 靳学慧. 黑龙江省稻瘟病菌无毒基因的检测[J]. 作物杂志, 2020, (3): 197–200 |
| [4] | 张晓玉,张亚玲,靳学慧,闫天雨,赵泽. 稻瘟病菌杂交后代致病性遗传分析[J]. 作物杂志, 2020, (2): 182–187 |
| [5] | 岳琳祺,施卫萍,郭佳晖,郭平毅,郭杰. 谷子角质合成基因对干旱胁迫的响应[J]. 作物杂志, 2019, (4): 183–190 |
| [6] | 罗海斌, 蒋胜理, 黄诚梅, 曹辉庆, 邓智年, 吴凯朝, 徐林, 陆珍, 魏源文. 甘蔗ScHAK10基因克隆及表达分析[J]. 作物杂志, 2018, (4): 53–61 |
| [7] | 朱畇昊,爼梦航,苏秀红,董诚明,陈随清. 冬凌草HMGS基因的克隆与表达分析[J]. 作物杂志, 2016, (5): 25–30 |
| [8] | 马军韬, 张国民, 辛爱华, 等. 以稻瘟病抗性基因分析为基础的水稻品种抗性布局研究[J]. 作物杂志, 2015, (1): 151–155 |
| [9] | 雷财林, 王久林, 程治军, 等. 水稻抗稻瘟病单基因鉴别体系研究的概况与展望[J]. 作物杂志, 2014, (2): 5–8 |
| [10] | 罗生香, 王倩, 张帆, 等. 水稻品种空育131的稻瘟菌致病菌株的遗传多样性分析[J]. 作物杂志, 2013, (4): 133–136 |
| [11] | 宋福金. 黑龙江省水稻稻瘟病大发生的原因分析与对策[J]. 作物杂志, 2006, (1): 69–69 |
| [12] | 雷财林, 凌忠专, 王久林, 等. 北方稻区稻瘟病菌生理小种变化与抗病育种策略[J]. 作物杂志, 2000, (3): 14–16 |
| [13] | 雷财林, 王久林, 凌忠专. 稻瘟病菌生理小种的变化动态[J]. 作物杂志, 1996, (4): 35–37 |
| [14] | 王东平. 略谈四川省杂交水稻的抗稻瘟病育种[J]. 作物杂志, 1995, (4): 30–31 |
| [15] | 凌忠专. 浅谈抗稻瘟病育种问题[J]. 作物杂志, 1989, (3): 35–36 |
|
||