作物杂志,2022, 第1期: 77–83 doi: 10.16035/j.issn.1001-7283.2022.01.011
苦荞FtC4H基因克隆与生物信息学分析
尹桂芳1( ), 段迎2, 杨晓琳2, 蔡苏云2, 王艳青1, 卢文洁1, 孙道旺1, 贺润丽2(
), 段迎2, 杨晓琳2, 蔡苏云2, 王艳青1, 卢文洁1, 孙道旺1, 贺润丽2( ), 王莉花1(
), 王莉花1( )
)
- 1云南省农业科学院生物技术与种质资源研究所/云南省农业生物技术重点实验室/农业农村部西南作物基因资源与种质创制重点实验室,650205,云南昆明
2山西中医药大学中药与食品工程学院,030619,山西太原
Cloning and Bioinformatics Analysis of FtC4H Gene from Tartary Buckwheat
Yin Guifang1( ), Duan Ying2, Yang Xiaolin2, Cai Suyun2, Wang Yanqing1, Lu Wenjie1, Sun Daowang1, He Runli2(
), Duan Ying2, Yang Xiaolin2, Cai Suyun2, Wang Yanqing1, Lu Wenjie1, Sun Daowang1, He Runli2( ), Wang Lihua1(
), Wang Lihua1( )
)
- 1Biotechnology and Germplasm Resources Institute, Yunnan Academy of Agricultural Sciences/ Yunnan Provincial Key Laboratory of Agricultural Biotechnology/Key Laboratory of Southwestern Crop Gene Resources and Germplasm Innovation, Ministry of Agriculture and Rural Affairs, Kunming 650205, Yunnan, China
2College of Traditional Chinese Medicine and Food Engineering, Shanxi University of Chinese Medicine, Taiyuan 030619, Shanxi, China
摘要:
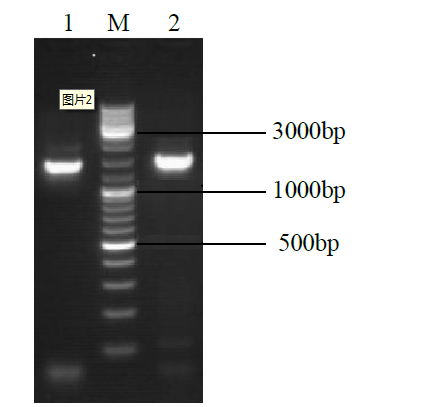
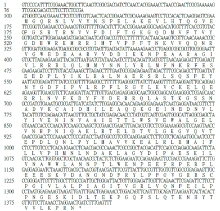
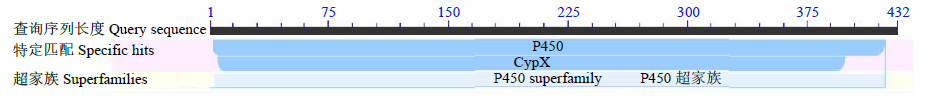
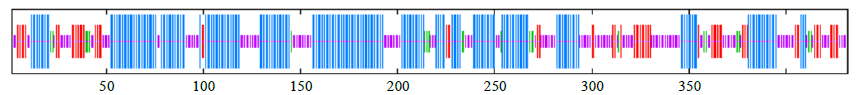
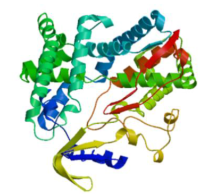
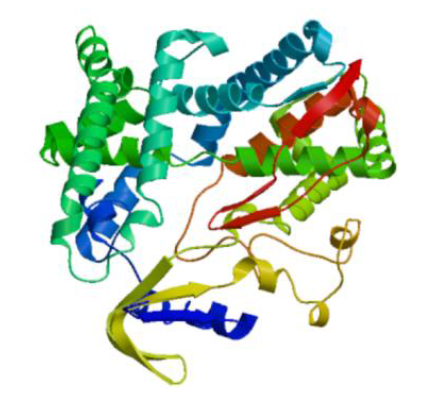
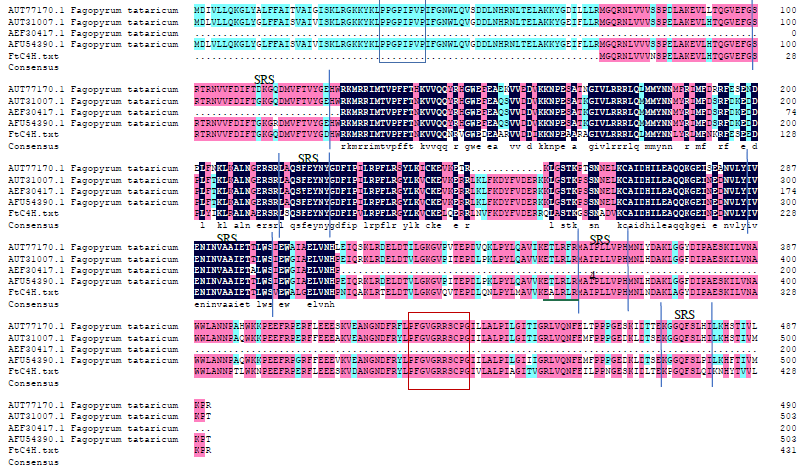
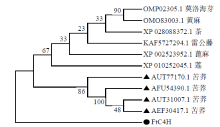
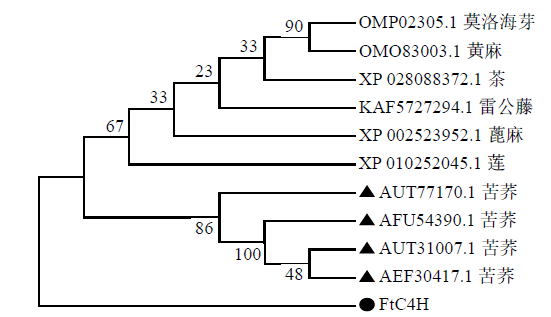
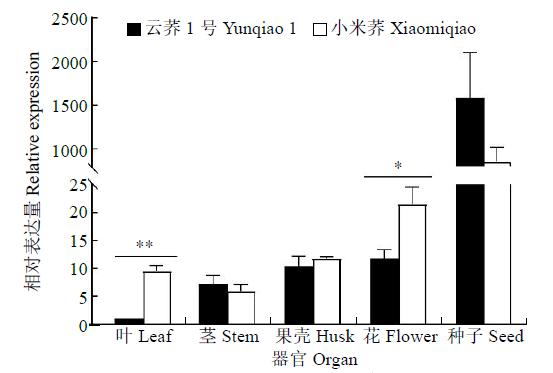
克隆苦荞苯丙烷类物质代谢途径中的关键酶肉桂酸-4-羟基化酶基因(FtC4H),为进一步研究其功能奠定基础。以云荞1号和小米荞为材料,提取不同发育期果壳RNA,利用RT-PCR法克隆苦荞FtC4H基因,运用生物信息学分析FtC4H蛋白的特征,构建FtC4H蛋白系统进化树,分析其基因表达。结果表明,克隆的FtC4H基因序列包含1299bp完整的cDNA开放阅读框,编码432个氨基酸,为亲水性不稳定碱性蛋白,具有P450超家族保守域,不具有跨膜结构域,有丰富的二级结构,三级结构预测显示FtC4H与6vby.1.A的序列相似度高。系统进化分析结果表明,本研究克隆的FtC4H与已报道的苦荞其他C4H基因不同。qRT-PCR结果表明,FtC4H在小米荞的花和叶中相对表达量显著高于云荞1号。
| [1] | 中国植物志编辑委员会. 中国植物志. 第25卷. 北京: 科学出版社, 1998. |
| [2] | 阮池银. 云南小凉山彝族苦荞文化的环境人类学研究. 昆明:云南大学, 2012. |
| [3] |
Bai C Z, Feng M L, Hao X L, et al. Rutin,quercetin,and free amino acid analysis in buckwheat (Fagopyrum) seeds from different locations. Genetics and Molecular Research, 2015, 14(4):19040-19048.
doi: 10.4238/2015.December.29.11 pmid: 26782554 |
| [4] |
Hu Y, Hou Z, Yi R, et al. Tartary buckwheat flavonoids ameliorate high fructose-induced insulin resistance and oxidative stress associated with the insulin signaling and Nrf2/HO-1 pathways in mice. Food and Function, 2017, 8(8):2803-2816.
doi: 10.1039/C7FO00359E |
| [5] |
Bao T, Wang Y, Li Y T, et al. Antioxidant and antidiabetic properties of tartary buckwheat rice flavonoids after in vitro digestion. Journal of Zhejiang University-Science B, 2016, 17(12):941-951.
doi: 10.1631/jzus.B1600243 |
| [6] |
Liu C L, Chen Y S, Yang J H, et al. Antioxidant activity of tartary (Fagopyrum tataricum (L.) Gaertn.) and common (Fagopyrum esculentum Moench) buckwheat sprouts. Journal of Agricultural and Food Chemistry, 2008, 56(1):173-178.
doi: 10.1021/jf072347s |
| [7] | 李玉英, 赵淑娟, 白崇智, 等. 苦荞异槲皮苷对人胃癌细胞SGC-7901增殖及凋亡的影响. 食品科学, 2014, 35(3):193-197. |
| [8] |
Hou Z X, Hu Y Y, Yang X B, et al. Antihypertensive effects of tartary buckwheat flavonoids by improvement of vascular insulin sensitivity in spontaneously hypertensive rats. Food and Function, 2017: 8(11):4217-4228.
doi: 10.1039/C7FO00975E |
| [9] |
Choi S Y, Choi J Y, Lee J M, et al. Tartary buckwheat on nitric oxide-induced inflammation in RAW264.7 macrophage cells. Food and Function, 2015, 6(8):2664-2670.
doi: 10.1039/C5FO00639B |
| [10] |
Russell D W, Conn E E. The cinnamic acid 4-hydroxylase of pea seedlings. Archives of Biochemistry and Biophysics, 1967, 122(1):256-258.
pmid: 4383827 |
| [11] |
Schilmiller A L, Stout J, Weng J K, et al. Mutations in the cinnamate 4-hydroxylase gene impact metabolism,growth and development in Arabidopsis. The Plant Journal, 2009, 60(5):771-782.
doi: 10.1007/s10725-019-00494-2 |
| [12] |
Park N I, Park J H, Park S U. Overexpression of cinnamate 4-hydroxylase gene enhances biosynthesis of decursinol angelate in Angelica gigas hairy roots. Molecular Biotechnology, 2012, 50(2):114-120.
doi: 10.1007/s12033-011-9420-8 pmid: 21626264 |
| [13] |
Singh K, Kumar S, Rani A, et al. Phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) and catechins (flavan-3-ols) accumulation in tea. Functional and Integrative Genomics, 2009, 9(1):125-134.
doi: 10.1007/s10142-008-0092-9 |
| [14] | 曾祥玲, 郑日如, 罗靖, 等. 桂花C4H基因的克隆与表达特性分析. 园艺学报, 2016, 43(3):525-537. |
| [15] | 程俊, 程曦, 盛玲玲, 等. 砀山酥梨肉桂酸4-羟化酶基因的克隆及表达分析. 农业生物技术学报, 2016, 24(11):1698-1708. |
| [16] |
Millar D J, Long M, Donovan G, et al. Introduction of sense constructs of cinnamate 4-hydroxylase (CYP73A24) in transgenic tomato plants shows opposite effects on flux into stem lignin and fruit flavonoids. Phytochemistry, 2007, 68(11):1497-1509.
pmid: 17509629 |
| [17] | Baek M H, Chung B Y, Kim J H, et al. cDNA cloning and expression pattern of Cinnamate-4-Hydroxylase in the Korean blackraspberry. Biochemistry and Molecular Biology Reports, 2008, 41(7):529-536. |
| [10] | Liu W, Zhu D, Liu D, et al. Comparative metabolic activity related to flavonoid synthesis in leaves and flowers of Chrysanthemum morifoliumin response to K deficiency. Plant and Soil, 2010, 335,325-337. |
| [18] |
Cheng S Y, Yan J P, Meng X X, et al. Characterization and expression patterns of a cinnamate-4-hydroxylase gene involved in lignin biosynthesis and in response to various stresses and hormonal treatments in Ginkgo biloba. Acta Physiologiae Plantarum, 2018, 40:1-15.
doi: 10.1007/s11738-017-2577-4 |
| [19] | 黄利娜, 吴光斌, 匡凤元, 等. 莲雾果实C4H基因的克隆及在NO处理下的表达分析. 集美大学学报(自然科学版), 2020, 25(2):105-112. |
| [20] | 陈鸿翰, 袁梦求, 李双江, 等. 苦荞肉桂酸羟化酶基因(FtC4H)的克隆及其UV-B胁迫下的组织表达. 农业生物技术学报, 2013, 21(2):137-147. |
| [21] | 刘荣华, 王丽, 孙朝霞, 等. 苦荞FtC4H基因的cDNA克隆及生物信息学分析. 山西农业大学学报(自然科学版), 2017, 37(11):767-773. |
| [22] | 王轶男, 陈雪, 盖颖. 毛白杨木质素合成酶基因F5H克隆与生物信息学分析. 广东农业科学, 2014, 41(20):131-135. |
| [23] |
Fahrendorf T, Dixon R A. Stress responses in alfalfa (Medicago sativa L.). XVIII:Molecular cloning and expression of the elicitor-inducible cinnamic acid 4-hydroxylase cytochrome P450. Archives of Biochemistry and Biophysics, 1993, 305:509-515.
pmid: 8373188 |
| [24] | Teutsch H G, Hasenfratz M P, Lesot A, et al. Isolation and sequence of a cDNA encoding the Jerusalem artichoke cinnamate 4-hydroxylase,a major plant cytochrome P450 involved in the general phenylpropanoid pathway. Proceedings of the National Academy of Sciences of the United States of America, 1993, 90(9):4102-4106. |
| [11] | Yan Q, Si J, Cui X X, et al. The soybean cinnamate 4-hydroxylase gene GmC4H1 contributes positively to plant defense via increasing lignin content. Plant Growth Regulation, 2019, 88(2):139-149. |
| [25] | 冯艺川, 赵洋, 全雪丽, 等. 膜荚黄芪C4H基因的克隆及表达分析. 分子植物育种, 2021, 19(1):130-136. |
| [1] | 翁文凤, 伍小方, 张凯旋, 唐宇, 江燕, 阮景军, 周美亮. 过表达FtbZIP5提高苦荞毛状根黄酮积累及其耐盐性[J]. 作物杂志, 2021, (4): 1–9 |
| [2] | 贾瑞玲, 赵小琴, 南铭, 陈富, 刘彦明, 魏立平, 刘军秀, 马宁. 64份苦荞种质资源农艺性状遗传多样性分析与综合评价[J]. 作物杂志, 2021, (3): 19–27 |
| [3] | 靳建刚, 田再芳. 山西北部地区引种苦荞品种的灰色关联度分析[J]. 作物杂志, 2021, (2): 52–56 |
| [4] | 马名川, 刘龙龙, 刘璋, 周建萍, 南成虎, 张丽君. 苦荞全基因组SSR位点特征分析与分子标记开发[J]. 作物杂志, 2021, (1): 38–46 |
| [5] | 卢晓玲, 何铭, 张凯旋, 廖志勇, 周美亮. 苦荞鼠李糖基转移酶FtF3GT1基因的克隆与转化毛状根研究[J]. 作物杂志, 2020, (5): 33–40 |
| [6] | 杨学乐, 张璐, 李志清, 何录秋. 苦荞种质资源表型性状的遗传多样性分析[J]. 作物杂志, 2020, (5): 53–58 |
| [7] | 李春花, 黄金亮, 尹桂芳, 王艳青, 卢文洁, 孙道旺, 王春龙, 郭来春, 洪波, 任长忠, 王莉花. 苦荞粒形相关性状的遗传分析[J]. 作物杂志, 2020, (3): 42–46 |
| [8] | 马成瑞,向达兵,万燕,欧阳建勇,宋月,唐正松,刘建英,赵钢. 不同苦荞品种花和籽粒空间分布特征及差异分析[J]. 作物杂志, 2020, (1): 35–40 |
| [9] | 杨甜,张永清,董馥慧,马星星,薛小娇. 不同水分条件下不同抗旱性苦荞根系生长规律研究[J]. 作物杂志, 2019, (6): 76–82 |
| [10] | 岳琳祺,施卫萍,郭佳晖,郭平毅,郭杰. 谷子角质合成基因对干旱胁迫的响应[J]. 作物杂志, 2019, (4): 183–190 |
| [11] | 宋丽芳,冯美臣,张美俊,肖璐洁,王超,杨武德,宋晓彦. 外源硒对苦荞生长发育及子粒硒含量的影响[J]. 作物杂志, 2019, (3): 150–154 |
| [12] | 马名川,刘龙龙,张丽君,崔林,周建萍. EMS诱变刺荞的形态突变体鉴定与分析[J]. 作物杂志, 2019, (3): 37–41 |
| [13] | 崔娅松, 王艳, 杨丽娟, 吴朝昕, 周飘, 冉盼, 陈庆富. 米苦荞果壳率及其相关性状的遗传研究[J]. 作物杂志, 2019, (2): 51–60 |
| [14] | 赵鑫,陈少锋,王慧,刘三才,杨修仕,张宝林. 晋北地区不同苦荞品种产量和品质研究[J]. 作物杂志, 2018, (5): 27–32 |
| [15] | 罗海斌, 蒋胜理, 黄诚梅, 曹辉庆, 邓智年, 吴凯朝, 徐林, 陆珍, 魏源文. 甘蔗ScHAK10基因克隆及表达分析[J]. 作物杂志, 2018, (4): 53–61 |
|
||