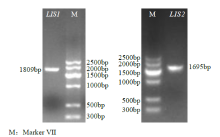
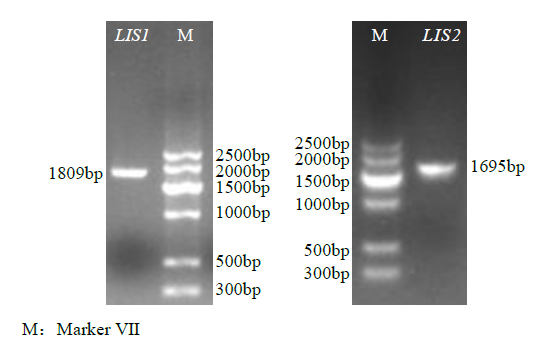
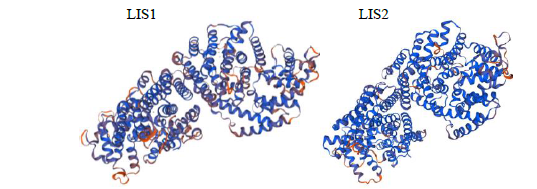
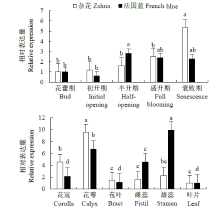
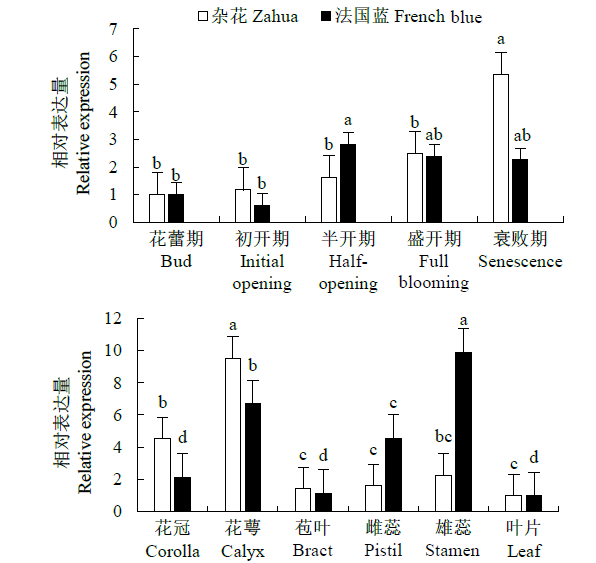
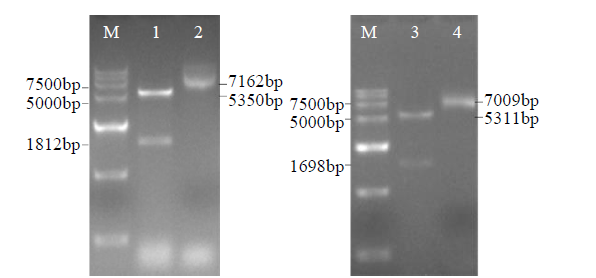
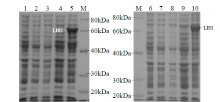
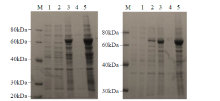
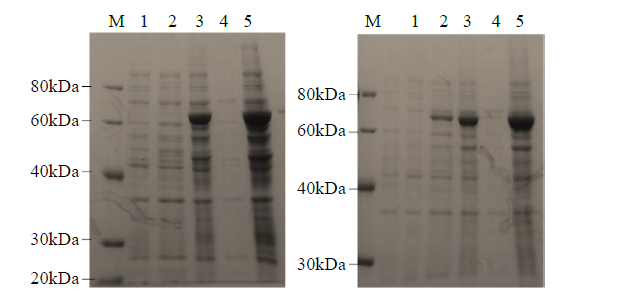
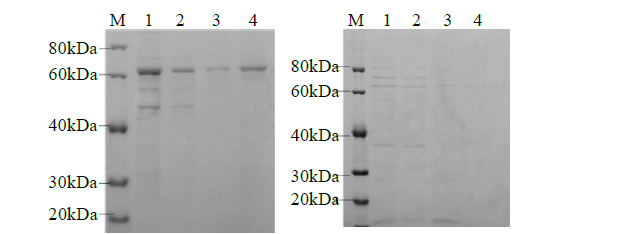
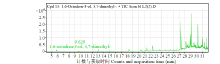
Crops ›› 2021, Vol. 37 ›› Issue (6): 78-87.doi: 10.16035/j.issn.1001-7283.2021.06.013
Previous Articles Next Articles
Cloning, Expression and Enzyme Activity Detection of Linalool Synthase Gene in Lavender
Gong Lintao( ), Su Xiujuan(
), Su Xiujuan( ), Liao Yan, Keremuhan·Wusiman , Zhou Di, Yin Songsong
), Liao Yan, Keremuhan·Wusiman , Zhou Di, Yin Songsong
- College of Agriculture, Xinjiang Agricultural University/Key Laboratory of Agricultural Biotechnology, Xinjiang Agricultural University, Urumqi 830052, Xinjiang, China
| [1] | 高婷婷, 谭勇, 魏雯, 等. 薰衣草的研究进展. 海峡两岸暨CSNR全国中药及天然药物资源学术研讨会, 2012:737-740. |
| [2] | 王玉芹, 孙亚军, 施献儿. 薰衣草精油的化学成分与药理活性. 国外医药(植物药分册), 2004(1):5-8. |
| [3] |
Woronuk G, Demissie Z, Rheault M, et al. Biosynthesis and therapeutic properties of Lavandula essential oil constituents. Planta Medica, 2011, 77(1):7-15.
doi: 10.1055/s-0030-1250136 pmid: 20665367 |
| [4] | 樊荣辉, 黄敏玲, 钟淮钦, 等. 花香的生物合成、调控及基因工程研究进展. 中国细胞生物学报, 2011, 33(9):1028-1036. |
| [5] | 陈尚钘, 赵玲华, 徐小军. 天然芳樟醇资源及其开发利用. 林业工程学报, 2013, 27(2):13-17. |
| [6] |
Kessler A, Baldwin I T. Defensive function of herbivore-induced plant volatile emissions in nature. Science, 2001, 291(5511):2141-2144.
pmid: 11251117 |
| [7] | 刘静, 徐江涛. 薰衣草精油对小鼠镇静催眠作用的实验研究. 临床和实验医学杂志, 2012, 11(18):1440-1441. |
| [8] |
Linck V D M, Silva A L D, Micheli Figueiró, et al. Inhaled linalool-induced sedation in mice. Phytomedicine, 2009, 16(4):303-307.
doi: 10.1016/j.phymed.2008.08.001 |
| [9] |
Pichersky E, Lewinsohn E, Croteau R. Purification and characterization of S-linalool synthase,an enzyme involved in the production of floral scent in Clarkia breweri. Archives of Biochemistry and Biophysics, 1995, 316(2):803-807.
pmid: 7864636 |
| [10] |
Dudareva N, Cseke L, Blanc V M, et al. Evolution of floral scent in Clarkia:novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell, 1996, 8(7):1137-1148.
pmid: 8768373 |
| [11] |
Mendoza-Poudereux I, Muñoz-Bertomeu J, Navarro A, et al. Enhanced levels of S-linalool by metabolic engineering of the terpenoid pathway in spike lavender leaves. Metabolic Engineering, 2014, 23:136-144.
doi: 10.1016/j.ymben.2014.03.003 pmid: 24685653 |
| [12] |
Lewinsohn E, Schalechet F, Wilkinson J, et al. Enhanced levels of the aroma and flavor compound S-linalool by metabolic engineering of the terpenoid pathway in tomato fruits. Plant Physiology, 2001, 127(3):1256-1265.
pmid: 11706204 |
| [13] | He J, Fandino R A, Halitschke R, et al. An unbiased approach elucidates variation in (S)-(+)-linalool,a context-specific mediator of a tri-trophic interaction in wild tobacco. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(29):14651-14660. |
| [14] |
Tengxun Z, Guo Y H, Shi X J, et al. Overexpression of LiTPS2 from a cultivar of lily (Lilium ‘Siberia’) enhances the monoterpenoids content in tobacco flowers. Plant Physiology and Biochemistry, 2020, 151:391-399.
doi: S0981-9428(20)30161-3 pmid: 32278293 |
| [15] | Zeng X L, Liu C, Zheng R R, et al. Emission and accumulation of monoterpene and the key terpene synthase (TPS) associated with monoterpene biosynthesis in Osmanthus fragrans Lour. Frontiers in Plant Science, 2015, 6:1232. |
| [16] | 张雪荣. 薰衣草芳樟醇合酶的基因克隆、功能鉴定及转基因技术的研究. 呼和浩特:内蒙古农业大学, 2007. |
| [17] | 赵钟鑫, 王健, 李琴, 等. 阔叶薰衣草芳樟醇合酶基因的克隆与表达载体构建. 植物研究, 2013, 33(3):308-316. |
| [18] | 方柳. 莽山野柑特征香气相关候选基因克隆和功能鉴定. 武汉:华中农业大学, 2014. |
| [19] |
Huang X Z, Xiao Y T, KöLlner T G, et al. The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell and Environment, 2018, 41(1):261-274.
doi: 10.1111/pce.v41.1 |
| [20] |
Cseke L, Dudareva N, Pichersky E. Structure and evolution of linalool synthase. Molecular Biology and Evolution, 1998, 15(11):1491-1498.
pmid: 12572612 |
| [21] |
Raguso R A, Pichersky E. New perspectives in pollination biology:floral fragrances. A day in the life of a linalool molecule:chemical communication in a plant-pollinator system. Part 1:linalool biosynthesis in flowering plants. Plant Species Biology, 1999, 14(2):95-120.
doi: 10.1046/j.1442-1984.1999.00014.x |
| [22] |
Yu F, Utsumi R. Diversity,regulation,and genetic manipulation of plant mono- and sesquiterpenoid biosynthesis. Cellular and Molecular Life Sciences, 2009, 66(18):3043-3052.
doi: 10.1007/s00018-009-0066-7 |
| [23] |
Lin Y L, Lee Y R, Huang W K, et al. Characterization of S-(+)-linalool synthase from several provenances of Cinnamomum osmophloeum. Tree Genetics and Genomes, 2014, 10(1):75-86.
doi: 10.1007/s11295-013-0665-1 |
| [24] |
Yue Y, Yu R, Fan Y. Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium. Planta, 2014, 240(4):745-762.
doi: 10.1007/s00425-014-2127-x |
| [25] |
Gao F Z, Liu B F, Li M, et al. Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia x hybrida. Journal of Experimental Botany, 2018, 69(18):4249-4265.
doi: 10.1093/jxb/ery224 |
| [26] |
Jiang Y, Qian R, Zhang W, et al. Composition and biosynthesis of scent compounds from sterile flowers of an ornamental plant Clematis florida cv. ‘Kaiser’. Molecules, 2020, 25(7):1711.
doi: 10.3390/molecules25071711 |
| [27] |
Fathi E, Majdi M, Dastan D, et al. The spatio-temporal expression of some genes involved in the biosynthetic pathways of terpenes/phenylpropanoids in yarrow (Achillea millefolium). Plant Physiology and Biochemistry, 2019, 142:43-52.
doi: 10.1016/j.plaphy.2019.06.036 |
| [28] |
Biswas K K, Foster A J, Aung T, et al. Essential oil production:relationship with abundance of glandular trichomes in aerial surface of plants. Acta Physiologiae Plantarum, 2009, 31(1):13-19.
doi: 10.1007/s11738-008-0214-y |
| [29] |
Chuang L, Wen C H, Lee Y R, et al. Identification,functional characterization,and seasonal expression patterns of five sesquiterpene synthases in Liquidambar formosana. Journal of Natural Products, 2018, 81(5):1162-1172.
doi: 10.1021/acs.jnatprod.7b00773 pmid: 29746128 |
| [30] |
Magnard J L, Aurélie R B, Bettini F, et al. Linalool and linalool nerolidol synthases in roses,several genes for little scent. Plant Physiology and Biochemistry, 2018, 127:74-87.
doi: 10.1016/j.plaphy.2018.03.009 |
| [31] |
Hattan J, Shindo K, Ito T, et al. Identification of a novel hedycaryol synthase gene isolated from Camellia brevistyla flowers and floral scent of Camellia cultivars. Planta, 2016, 243(4):959-972.
doi: 10.1007/s00425-015-2454-6 |
| [32] |
Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Molecular Biology and Evolution, 1985, 2(1):13-34.
pmid: 3916708 |
| [33] | 刘琬菁, 吕海舟, 李滢, 等. 植物萜类合酶研究新进展. 植物生理学报, 2017, 53(7):1139-1149. |
| [1] | Song Kunfeng, Hao Fengsheng, Yao Xin, Wang Jing, Liu Weiqun. Cloning and Functional Analysis of Tobacco Nt14-3-3-like C Gene [J]. Crops, 2021, 37(5): 6-13. |
| [2] | Wang Tong, Zhao Xiaodong, Zhen Pingping, Chen Jing, Chen Mingna, Chen Na, Pan Lijuan, Wang Mian, Xu Jing, Yu Shanlin, Chi Xiaoyuan, Zhang Jiancheng. Genome-Wide Identification and Characteristic Analyzation of the TCP Transcription Factors Family in Peanut [J]. Crops, 2021, 37(2): 35-44. |
| [3] | Liu Xiaoli, Han Litao, Wei Nan, Shen Fei, Cai Yilin. Cloning, Molecular Characteristics Analysis and Genetic Transformation of Arabidopsis thaliana of ZmGS5 Gene in Maize [J]. Crops, 2021, 37(1): 16-25. |
| [4] | Lu Xiaoling, He Ming, Zhang Kaixuan, Liao Zhiyong, Zhou Meiliang. Study on the Cloning and Transformation of Rhamnose Transferase FtF3GT1 Gene in Tartary Buckwheat [J]. Crops, 2020, 36(5): 33-40. |
| [5] | Haibin Luo, Shengli Jiang, Chengmei Huang, Huiqing Cao, Zhinian Deng, Kaichao Wu, Lin Xu, Zhen Lu, Yuanwen Wei. Cloning and Expression of ScHAK10 Gene in Sugarcane [J]. Crops, 2018, 34(4): 53-61. |
| [6] | Xuemei Li,Chang Liu,Qianwen Liu,Yudi Zhang,Xuemei Li. The Effects of PEG Pretreatment on the Isozymes and Expression of Antioxidant Enzymes in Rice Leaves under Water Stress [J]. Crops, 2016, 32(6): 107-111. |
| [7] | Zanping Han,Yanhui Chen,Shulei Guo,Xiaofeng Zu,Shunxi Wang,Xiyong Zhao. Cloning and Function Analysis of Resistance Gene ZmqLTG3-1 in Maize [J]. Crops, 2016, 32(4): 47-55. |
| [8] | Yu Li,Wenyin Zheng,Chun Feng,Rongfu Wang,Juan Li. Analysis of FeSOD Gene Expression in Normal Wheat Yannong 19 Seedlings under Abiotic Stress [J]. Crops, 2016, 32(4): 75-79. |
| [9] | Hongmei Yuan,Wendong Guo,Lijuan Zhao,Ying Yu,Jianzhong Wu,Lili Cheng,Dongsheng Zhao,Qinghua Kang,Wengong Huang,Yubo Yao,Xixia Song,Weidong Jiang,Yan Liu,Tingfen Ma,Guangwen Wu,Fengzhi Guan. Cloning and Expression Analysis of the Glycosyltransferase Gene LuUGT72E1 in Flax [J]. Crops, 2016, 32(4): 62-67. |
|
||