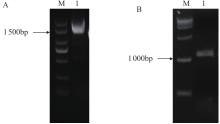
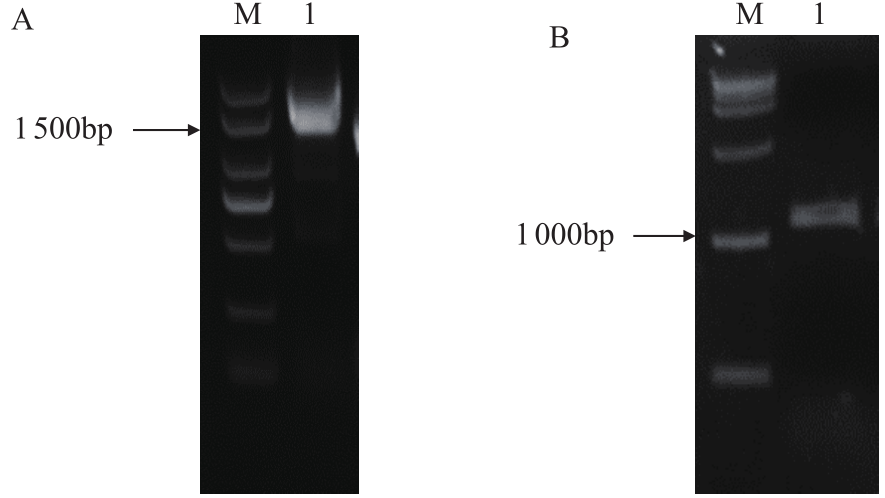
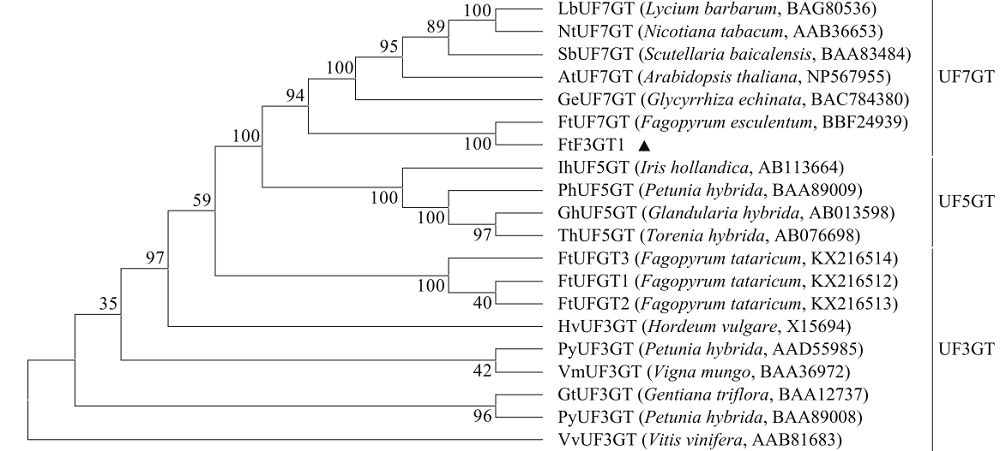
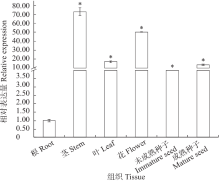
Crops ›› 2020, Vol. 36 ›› Issue (5): 33-40.doi: 10.16035/j.issn.1001-7283.2020.05.005
Previous Articles Next Articles
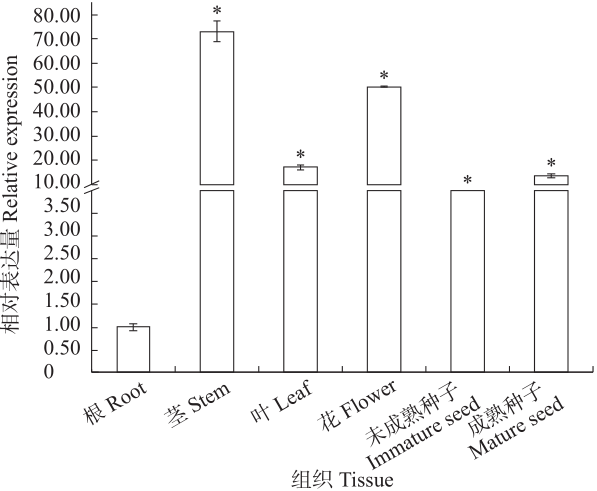
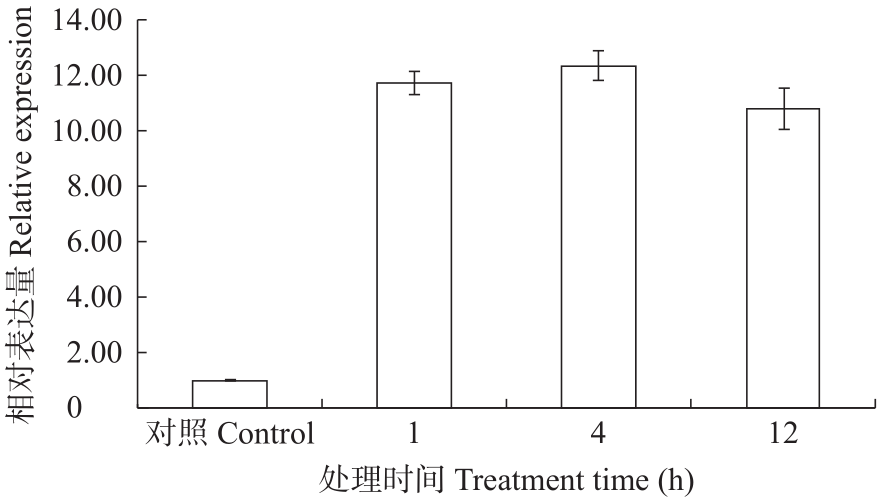
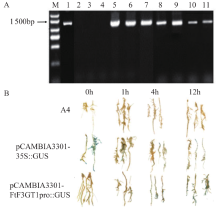
Study on the Cloning and Transformation of Rhamnose Transferase FtF3GT1 Gene in Tartary Buckwheat
Lu Xiaoling1,2( ), He Ming2, Zhang Kaixuan2, Liao Zhiyong1(
), He Ming2, Zhang Kaixuan2, Liao Zhiyong1( ), Zhou Meiliang2(
), Zhou Meiliang2( )
)
- 1College of Life and Environmental Science, Wenzhou University, Wenzhou 325035, Zhejiang, China
2Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
| [1] |
Wu J C, Lai C S, Tsai M L, et al. Chemopreventive effect of natural dietary compounds on xenobiotic-induced toxicity. Journal of Food and Drug Analysis, 2017,25(1):176-186.
pmid: 28911535 |
| [2] |
Moon Y J, Wang X, Morris M E. Dietary flavonoids:Effects on xenobiotic and carcinogen metabolism. Toxicology in Vitro, 2006,20(2):187-210.
doi: 10.1016/j.tiv.2005.06.048 pmid: 16289744 |
| [3] |
Cushnie T P T, Lamb A J. Antimicrobial activity of flavonoids. International Journal of Antimicrobial Agents, 2005,26(5):343-356.
doi: 10.1016/j.ijantimicag.2005.09.002 pmid: 16323269 |
| [4] |
Cushnie T P, Lamb A J. Recent advances in understanding the antibacterial properties of flavonoids. International Journal of Antimicrobial Agents, 2011,38(2):99-107.
doi: 10.1016/j.ijantimicag.2011.02.014 |
| [5] |
Buer C S, Imin N, Djordjevic M A. Flavonoids:new roles for old molecules. Journal of Integrative Plant Biology, 2010,52(1):98-111.
doi: 10.1111/j.1744-7909.2010.00905.x |
| [6] | 范昱, 丁梦琦, 张凯旋, 等. 荞麦种质资源概况. 植物遗传资源学报, 2019,20(4):813-828. |
| [7] |
Bai C Z, Feng M L, Hao X L, et al. Rutin,quercetin,and free amino acid analysis in buckwheat (Fagopyrum) seeds from different locations. Genetics and Molecular Research, 2015,14(4):19040-19048.
doi: 10.4238/2015.December.29.11 pmid: 26782554 |
| [8] | 李蒙. 荞麦生物活性肽的研究进展. 江西农业学报, 2018,30(12):17-21. |
| [9] |
Zhou M L, Sun Z M, Ding M Q, et al. FtSAD2 and FtJAZ1 regulate activity of the FtMYB11 transcription repressor of the phenylpropanoid pathway in Fagopyrum tataricum. New Phytologist, 2017,216(3):814-828.
doi: 10.1111/nph.14692 pmid: 28722263 |
| [10] |
Zhou M L, Memelink J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnology Advances, 2016,34(4):441-449.
doi: 10.1016/j.biotechadv.2016.02.004 pmid: 26876016 |
| [11] |
An X H, Tian Y, Chen K Q, et al. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant & Cell Physiology, 2015,56(4):650-662.
doi: 10.1093/pcp/pcu205 pmid: 25527830 |
| [12] |
Gonzalez A, Zhao M, Leavitt J M, et al. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal, 2008,53(5):814-827.
doi: 10.1111/j.1365-313X.2007.03373.x pmid: 18036197 |
| [13] |
Boter M, Golz J F, Gimenez-Ibanez S, et al. Filamentous flower is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell, 2015,27(11):3160-3174.
doi: 10.1105/tpc.15.00220 pmid: 26530088 |
| [14] |
Lim S H, Kim D H, Kim J K, et al. A radish basic helix-loop-helix transcription factor,RsTT8 acts a positive regulator for anthocyanin biosynthesis. Frontiers in Plant Science, 2017,8:1917-1931.
doi: 10.3389/fpls.2017.01917 pmid: 29167678 |
| [15] |
Tohge T, Watanabe M, Hoefgen R, et al. The evolution of phenylpropanoid metabolism in the green lineage. Critical Reviews in Biochemistry and Molecular Biology, 2013,48(2):123-152.
doi: 10.3109/10409238.2012.758083 |
| [16] |
He F, Mu L, Yan G L, et al. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules, 2010,15(12):9057-9091.
doi: 10.3390/molecules15129057 pmid: 21150825 |
| [17] | Du H, Huang Y B, Tang Y X. Genetic and metabolic engineering of isoflavonoid biosynthesis. Applied Microbiology and Biotech-nology, 2010,86(5):1293-1312. |
| [18] | Rio D C, Ares M, Hannon G J, et al. Purification of RNA using trizol. Cold Spring Harbor Protocols, 2010,(6):1-3. |
| [19] | 谈天斌, 卢晓玲, 张凯旋, 等. TrMYB308基因的克隆及在苦荞毛状根中的功能分析. 植物遗传资源学报, 2019,20(6):1542-1553. |
| [20] |
Zhou M L, Zhu X M, Shao J R, et al. Transcriptional response of the catharanthine biosynthesis pathway to methyl jasmonate/nitric oxide elicitation in Catharanthus roseus hairy root culture. Applied Microbiology and Biotechnology, 2010,88(3):737-750.
doi: 10.1007/s00253-010-2822-x |
| [21] | 李为喜. AlCl3分光光度法测定荞麦种质资源中黄酮的研究. 植物遗传资源学报, 2008,9(4):502-505. |
| [22] |
Nakatsuka T, Sato K, Takahashi H, et al. Cloning and characterization of the UDP-glucose:anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian. Journal of Experimental Botany, 2008,59(6):1241-1252.
doi: 10.1093/jxb/ern031 pmid: 18375606 |
| [23] |
Hsu Y H, Tagami T, Matsunaga K, et al. Functional characterization of UDP-rhamnose-dependent rhamnosyltransferase involved in anthocyanin modification,a key enzyme determining blue coloration in Lobelia erinus. The Plant Journal, 2017,89(2):325-337.
doi: 10.1111/tpj.13387 pmid: 27696560 |
| [24] |
Miller K D, Guyon V, Evans J N S, et al. Purification,cloning,and heterologous expression of a catalytically efficient flavonol 3-O-galactosyltransferase expressed in the male gametophyte of Petunia hybrida. The Journal of Biological Chemistry, 1999,274(48):34011-34019.
doi: 10.1074/jbc.274.48.34011 pmid: 10567367 |
| [25] |
Li X J, Zhang J Q, Wu Z C, et al. Functional characterization of a glucosyltransferase gene,LcUFGT1,involved in the formation of cyanidin glucoside in the pericarp of Litchi chinensis. Physiologia Plantarum, 2016,156(2):139-149.
doi: 10.1111/ppl.12391 pmid: 26419221 |
| [26] | Campell J A, Davies G J, Bulone V, et al. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochemistry, 1998,329(3):719. |
| [27] | 胡滢滢, 薛飞燕, 杨明峰, 等. 高山红景天UGT72B14G2基因克隆及其原核表达. 北京农学院学报, 2016,31(4):1-5. |
| [28] |
Koja E, Ohata S, Maruyama Y, et al. Identification and characterization of a rhamnosyltransferase involved in rutin biosynthesis in Fagopyrum esculentum (common buckwheat). Bioscience,Biotechnology, and Biochemistry, 2018,82(10):1790-1802.
doi: 10.1080/09168451.2018.1491286 pmid: 29972345 |
| [29] |
Perez A C, Goossens A. Jasmonate signalling:a copycat of auxin signalling? Plant, Cell and Environment, 2013,36(12):2071-2084.
doi: 10.1111/pce.12121 pmid: 23611666 |
| [30] |
Memelink J, Verpoorte R, Jan W K. ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends in Plant Science, 2001,6(5):212-219.
doi: 10.1016/s1360-1385(01)01924-0 pmid: 11335174 |
| [31] |
Misra P, Pandey A, Tiwari M, et al. Modulation of transcriptome and metabolome of tobacco by Arabidopsis transcription factor,AtMYB12,leads to insect resistance. Plant Physiology, 2010,152(4):2258-2268.
doi: 10.1104/pp.109.150979 pmid: 20190095 |
| [32] |
Zhang K, Logacheva M D, Meng Y, et al. Jasmonate-responsive MYB factors spatially repress rutin biosynthesis in Fagopyrum tataricum. Journal of Experimental Botany, 2018,69(8):1955-1966.
doi: 10.1093/jxb/ery032 pmid: 29394372 |
| [33] |
Zhang D, Jiang C L, Huang C H, et al. The light-induced transcription factor FtMYB116 promotes accumulation of rutin in Fagopyrum tataricum. Plant, Cell and Environment, 2018,42:1340-1351.
doi: 10.1111/pce.13470 pmid: 30375656 |
| [1] | Yang Xuele, Zhang Lu, Li Zhiqing, He Luqiu. Diversity Analysis of Tartary Buckwheat Germplasms Based on Phenotypic Traits [J]. Crops, 2020, 36(5): 53-58. |
| [2] | Li Chunhua, Huang Jinliang, Yin Guifang, Wang Yanqing, Lu Wenjie, Sun Daowang, Wang Chunlong, Guo Laichun, Hong Bo, Ren Changzhong, Wang Lihua. Genetic Analysis of Grain Shape Related Traits in Tartary Buckwheat [J]. Crops, 2020, 36(3): 42-46. |
| [3] | Chengrui Ma,Dabing Xiang,Yan Wan,Jianyong Ouyang,Yue Song,Zhengsong Tang,Jianying Liu,Gang Zhao. Difference Analysis of Spatial Distribution Characteristics of Different Tartary Buckwheat Varieties [J]. Crops, 2020, 36(1): 35-40. |
| [4] | Yang Tian,Zhang Yongqing,Dong Fuhui,Ma Xingxing,Xue Xiaojiao. Research on the Root Growth of Different Drought-Resistant Fagopyrum tataricum under Different Water Conditions [J]. Crops, 2019, 35(6): 76-82. |
| [5] | Song Lifang,Feng Meichen,Zhang Meijun,Xiao Lujie,Wang Chao,Yang Wude,Song Xiaoyan. Effects of Exogenous Selenium on the Growth and Development of Tartary Buckwheat and Selenium Content in Grains [J]. Crops, 2019, 35(3): 150-154. |
| [6] | Ma Mingchuan,Liu Longlong,Zhang Lijun,Cui Lin,Zhou Jianping. Morphological Identification and Analysis of EMS-Induced Mutants from Ciqiao [J]. Crops, 2019, 35(3): 37-41. |
| [7] | Yasong Cui, Yan Wang, Lijuan Yang, Chaoxin Wu, Piao Zhou, Pan Ran, Qingfu Chen. Genetic Analysis of Fruit Hull Rate and Related Traits on Tartary Buckwheat [J]. Crops, 2019, 35(2): 51-60. |
| [8] | Haibin Luo, Shengli Jiang, Chengmei Huang, Huiqing Cao, Zhinian Deng, Kaichao Wu, Lin Xu, Zhen Lu, Yuanwen Wei. Cloning and Expression of ScHAK10 Gene in Sugarcane [J]. Crops, 2018, 34(4): 53-61. |
| [9] | Yu Fan,Hongli Wang,Feng He,Dili Lai,Jiajun Wang,Yue Song,Dabing Xiang. Nutritional Quality in Seeds of Tartary Buckwheat Affected by After-Ripening [J]. Crops, 2018, 34(1): 96-101. |
| [10] | Yue Song,Dabing Xiang,Houbing Huang,Yu Fan,Shuang Wei,Sai Zhang. Lodging Resistance Identification and Evaluation of Different Tartary Buckwheat Cultivars [J]. Crops, 2017, 33(6): 65-71. |
| [11] | Xiushi Yang,Zhongxian Guo,Huimin Guo,Hui Wang,Sancai Liu. Effects of Sowing Date and Seeding Density on the Yield and Quality of Buckwheat [J]. Crops, 2017, 33(1): 88-93. |
| [12] | Zanping Han,Yanhui Chen,Shulei Guo,Xiaofeng Zu,Shunxi Wang,Xiyong Zhao. Cloning and Function Analysis of Resistance Gene ZmqLTG3-1 in Maize [J]. Crops, 2016, 32(4): 47-55. |
| [13] | Hongmei Yuan,Wendong Guo,Lijuan Zhao,Ying Yu,Jianzhong Wu,Lili Cheng,Dongsheng Zhao,Qinghua Kang,Wengong Huang,Yubo Yao,Xixia Song,Weidong Jiang,Yan Liu,Tingfen Ma,Guangwen Wu,Fengzhi Guan. Cloning and Expression Analysis of the Glycosyltransferase Gene LuUGT72E1 in Flax [J]. Crops, 2016, 32(4): 62-67. |
|
||