Crops ›› 2024, Vol. 40 ›› Issue (3): 82-89.doi: 10.16035/j.issn.1001-7283.2024.03.011
Previous Articles Next Articles
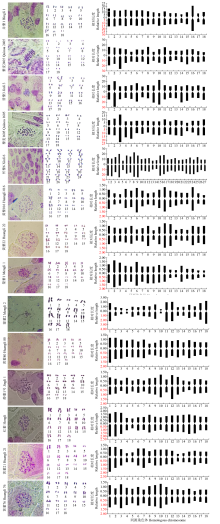
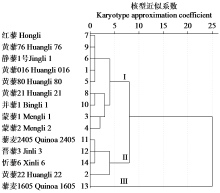
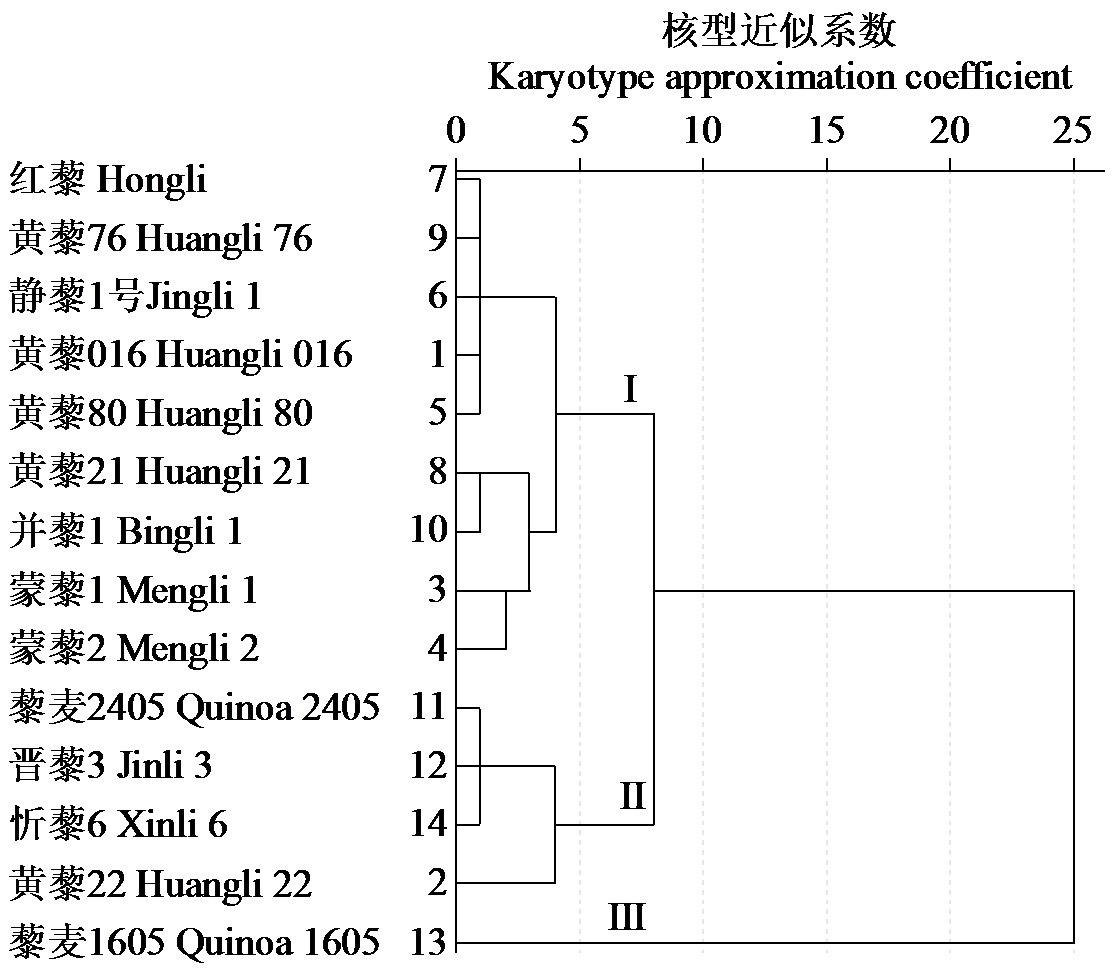
Chromosome Karyotype Analysis of 14 Quinoa Germplasms from Different Habitats
Liu Jianxia1,2( ), Wang Wenqing1, Xue Naiwen1,2, Guo Xuhu1,2, Ma Saiya1, Zhu Guofang1, Wen Riyu3(
), Wang Wenqing1, Xue Naiwen1,2, Guo Xuhu1,2, Ma Saiya1, Zhu Guofang1, Wen Riyu3( )
)
- 1Shanxi Datong University, Datong 037009, Shanxi, China
2Research and Development Center of Agricultural Facility Technology of Shanxi Datong University, Datong 037009, Shanxi, China
3Maize Research Institute, Shanxi Agricultural University, Xinzhou 034000, Shanxi, China
| [1] | 任贵兴, 叶全宝. 藜麦生产与应用. 北京: 科学出版社, 2013:49-50. |
| [2] | 陆红法, 张永正, 方美娟, 等. 浙江庆元高山藜麦营养成分分析. 浙江师范大学学报(自然科学版), 2017, 40(4):441-445. |
| [3] | Hong S Y, Cheon K S, Yoo K O, et al. Complete chloroplast genome sequences and comparative analysis of Chenopodium quinoa and C.album. Frontiers in Plant Science, 2017, 8:1696. |
| [4] | 权有娟, 李想, 袁飞敏, 等. 基于荧光原位杂交的藜属植物核型分析. 广西植物, 2021, 41(12):1988-1995. |
| [5] | 蒙昌智, 朱剑宏, 汪义勇. 食物新资源-南美藜. 四川省营养学会2000年学术会议专题报告及论文摘要汇编, 2000. |
| [6] | 王黎明, 马宁, 李颂, 等. 藜麦的营养价值及其应用前景. 食品工业科技, 2014, 35(1):381-384,389. |
| [7] |
胡一晨, 赵钢, 秦培友, 等. 藜麦活性成分研究进展. 作物学报, 2018, 44(11):1579-1591.
doi: 10.3724/SP.J.1006.2018.01579 |
| [8] |
Brittany L G, Patricio R S, Leonel E R. Innovations in health value and functional food development of quinoa (Chenopodium quinoa Willd.). Comprehensive Reviews in Food Science and Food Safety, 2015, 14:431-445.
pmid: 27453695 |
| [9] | 王艳青, 卢文洁, 李春花, 等. 10个藜麦新品系主要农艺性状分析与综合评价. 南方农业学报, 2019, 50(3):540-545. |
| [10] | 刘文瑜, 何斌, 杨发荣, 等. 不同品种藜麦幼苗对干旱胁迫和复水的生理响应. 草业科学, 2019, 36(10):2656-2666. |
| [11] |
杨发荣, 刘文瑜, 黄杰, 等. 河西地区2个藜麦品种引种试验研究. 草地学报, 2018, 26(5):1273-1276.
doi: 10.11733/j.issn.1007-0435.2018.05.034 |
| [12] | 魏玉明, 杨发荣, 刘文瑜, 等. 陇东旱塬区复种不同藜麦品种(系)的适应性初步评价. 西北农业学报, 2020, 29(5):675- 686. |
| [13] |
陆敏佳, 蒋玉蓉, 陆国权, 等. 利用SSR标记分析藜麦品种的遗传多样性. 核农学报, 2015, 29(2):260-269.
doi: 10.11869/j.issn.100-8551.2015.02.0260 |
| [14] | 权有娟, 刘博, 袁飞敏, 等. 不同籽粒颜色藜麦品种的核型分析. 华北农学报, 2020, 35(增1):72-77. |
| [15] | 雷海英, 侯沁文, 白凤麟, 等. 八种不同产地苦参的染色体数目及核型分析. 植物生理学报, 2019, 55(7):967-974. |
| [16] | Kawatani T, Ohno T.Chromosome numbers of genus Chenopodium I. Japanese Journal of Genetics, 1950, 25:177-180. |
| [17] | Kawatani T, Ohno T. Chromosome numbers of genus Chenopodium II. Japanese Journal of Genetics, 1956, 31:15-17. |
| [18] | Palomino G, Hemandez L T, Torres E D. Nuclear genome size and chromosome analysis in Chenopodium quinoa and C. berlandieri subsp. nuttalliae. Euphytica, 2008, 164:221-230. |
| [19] | Bhargava A, Shukia S, Ohri D. Karyotypic studies on some cultivated and wild species of Chenopodium (Chenopodiaceae). Genetic Resources and Crop Evolution, 2006, 53:1309-1320. |
| [20] | Kolano B, Tomczak H, Molewska R, et al. Distribution of 5S and 35S rRNA gene sites in 34 Chenopodium species (Amaranthaceae). Botanical Journal of the Linnean Society, 2012, 170:220-231. |
| [21] | 何燕, 邓永辉, 李梦寒, 等. 藜麦品系的染色体数目及核型分析. 西南大学学报(自然科学版), 2019, 41(1):27-31. |
| [22] | 乔永刚, 宋芸. 利用EXCEL制作核型模式图. 农业网络信息, 2006(10):97-98. |
| [23] | 李懋学, 陈瑞阳. 关于植物核型分析的标准化问题. 武汉植物学研究, 1985(4):297-302. |
| [24] |
乔永刚, 王勇飞, 曹亚萍, 等. 13种蒲公英属植物核型似近系数聚类分析. 草地学报, 2020, 28(1):285-290.
doi: 10.11733/j.issn.1007-0435.2020.01.035 |
| [25] | 杨光穗, 冷青云, 王呈丹, 等. 16个红掌品种的核型分析. 热带作物学报, 2016, 37(12):2283-2287. |
| [26] |
Ricroch A, Yockteng R, Brown S C, et al. Evolution of genome size across some cultivated Allium species. Genome, 2005, 48 (3):511-520.
pmid: 16121247 |
| [27] | Bhargava A, Rana T S, Shukla S, et al. Seed protein electrophoresis of some cultivated and wild species of Chenopodium. Biologia Plantarum, 2005, 49(4),505-511. |
| [28] | 夏雪, 田玉肖, 王祉琪, 等. 基于核型分析对芥蓝分类地位的探究. 分子植物育种, 2019, 17(7):2291-2296. |
| [29] | 闫素丽, 安玉麟, 孙瑞芬. 内葵杂3号染色体核型分析. 植物遗传资源学报, 2012, 11(6):784-788. |
| [30] |
卫尊征, 殷选红, 熊敏, 等. 3个彩色马蹄莲引进品种的核型分析. 植物遗传资源学报, 2012, 13(4):650-654.
doi: 10.13430/j.cnki.jpgr.2012.04.025 |
| [31] |
Wilson H, Manhart J. Crop/weed gene flow Chenopodium quinoa Willd. and C.berlandieri Moq. Theoretical and Applied Genetics, 1993, 86(5):642-648.
doi: 10.1007/BF00838721 pmid: 24193715 |
| [32] | Barker R E, Kilgore J A, Cook R L, et al. Use of flow cytometry to determine ploidy level of ryegrass. Seed Science and Technology, 2001, 29:493-502. |
| [33] | Kamemoto H, Shindo K, Kosoki K, et al. Chromosome homology in the Ceratobium, Phalaenanthe, and Latourea sections of the genus dendrobium. Pacific Science, 1964, 18:104-115. |
| [34] | Yan J, Zhang J, Sun K, et al. Ploidy level and DNA content of erianthus arundinaceus as determined by flow cytometry and the association with biological characteristics. PLoS ONE, 2016, 11(3):e0151948 |
| [35] |
任丽娟, 赵连生, 陈雅坤, 等. 基于主成分分析和聚类分析方法综合评价东北地区不同品种全株玉米青贮饲料的青贮品质. 动物营养学报, 2020, 32(8):3856-3868.
doi: 10.3969/j.issn.1006-267x.2020.08.044 |
| [1] | He Jiamin, Zhang Yongqing, Zhang Meng, Liang Ping, Wang Dan, Yan Fanfan. Effects of Seed Soaking with Uniconazole on Agronomic and Physiological Characteristics of Quinoa under Saline-Alkali Stress [J]. Crops, 2024, 40(2): 234-241. |
| [2] | Yang Enze, Wang Shuyan, Liu Ruixiang, Shi Fengyuan, Zhang Jinhao, Li Jiana, Li Zhiwei, Guo Zhanbin. Genetic Diversity Analysis of Quinoa Germplasm Resources Based on SRAP [J]. Crops, 2023, 39(6): 79-85. |
| [3] | Li Xinghe, Wang Haitao, Liu Cunjing, Tang Liyuan, Zhang Sujun, Cai Xiao, Zhang Xiangyun, Zhang Jianhong. QTL Mapping for Fiber Quality Traits Using Gossypium barbadense Chromosome Segment Introgression Lines [J]. Crops, 2023, 39(5): 1-9. |
| [4] | Guo Hongxia, Wang Chuangyun, Deng Yan, Zhao Li, Zhang Liguang, Guo Hongxia, Qin Lixia, Gao Fei, Xi Ruizhen. Response of Quinoa to Low Nitrogen Stress [J]. Crops, 2023, 39(3): 221-229. |
| [5] | Chen Cuiping, Yan Dianhai, Zhang Shumiao, Zuo Haonan, Gao Sen, Liu Yang. Fingerprint Construction and Genetic Diversity Analysis of Quinoa Based on SSR Markers [J]. Crops, 2023, 39(3): 35-42. |
| [6] | Liang Ping, Zhang Yongqing, Zhang Meng, Xue Xiaojiao, Li Pingping, Zhang Wenyan, Wang Dan, Zhao Gang. Effects of PAM Application Depth on the Growth and Physiological Indexes of Quinoa under Saline Alkali Stress [J]. Crops, 2023, 39(2): 178-185. |
| [7] | Mei Li. Research Progress and Development Prospect of Adaptive Cultivation of Quinoa in Beijing [J]. Crops, 2022, 38(6): 14-22. |
| [8] | Wang Siyu, Zuo Wenbo, Zhu Kaili, Guo Huimin, Xing Bao, Guo Yuqing, Bao Yuying, Yang Xiushi, Ren Guixing. Analysis and Evaluation of Agronomic Characteristics and Nutritional Qualities of 71 Quinoa Accessions [J]. Crops, 2022, 38(3): 63-72. |
| [9] | Qi Xiaolei, Li Xingfeng, Lü Guangde, Wang Ruixia, Wang Jun, Sun Xianyin, Sun Yingying, Chen Yongjun, Qian Zhaoguo, Wu Ke. Genetic Analysis of Taishan/Taikemai Serial Wheat Based on SNP Molecular Markers [J]. Crops, 2021, 37(5): 64-71. |
| [10] | Li Exian, Yin Fuyou, Zhang Dunyu, Chen Yue, Yu Tengqiong, Lei Yongtao, Xiao Suqin, Cheng Zaiquan, Ke Xue. Extraction of High-Quality Chromosome DNA in Yunnan Wild Rice Oryza officinalis [J]. Crops, 2020, 36(5): 103-109. |
| [11] | Wang Zhongqiu, Ying Pengfei, Chen Mengtao, He Qiongying, Hu Xin. Analysis of Grain and Quality Traits of Chromosome Arm Substitution Lines of Triticum dicoccoides in the Background of Triticum aestivum [J]. Crops, 2020, 36(4): 37-44. |
| [12] | Wang Yanqing,Li Yongjun,Li Chunhua,Lu Wenjie,Sun Daowang,Yin Guifang,Hong Bo,Wang Lihua. Correlation and Path Analysis of the Main Agronomic Traits and Yield per Plant of Quinoa [J]. Crops, 2019, 35(6): 156-161. |
| [13] | Haiyan Ding,Chunlin Wang,Yan Wu,Guoling Ren,Zuhuan Zhai,Jun Ma. Karyotype Analysis of Chromosome of Sunflower Longkuiza10 [J]. Crops, 2019, 35(2): 99-102. |
| [14] | Hongliang Cui,Bao Xing,Qing Yao,Qinping Zhang,Xiushi Yang,Yang Yao,Guixing Ren,Peiyou Qin. SWOT Analysis on Development of Quinoa Industry in Ili Valley of Xinjiang [J]. Crops, 2019, 35(1): 32-37. |
| [15] | Riyu Wen,Jianxia Liu,Zhenhua Zhang,Yaodong Guo,Xuyao Dai,Qingguo Jiang,Lisheng Fan. Effects of Drought Stress on Germination and Physiological Characteristics of Different Quinoa Seeds [J]. Crops, 2019, 35(1): 121-126. |
|
||