Crops ›› 2021, Vol. 37 ›› Issue (4): 10-17.doi: 10.16035/j.issn.1001-7283.2021.04.002
Previous Articles Next Articles
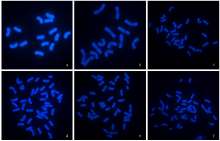
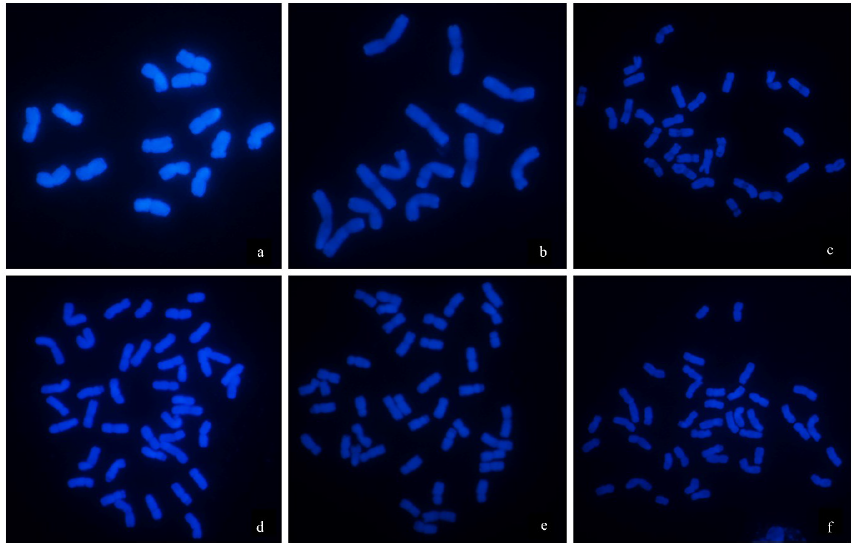
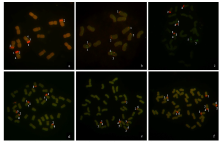
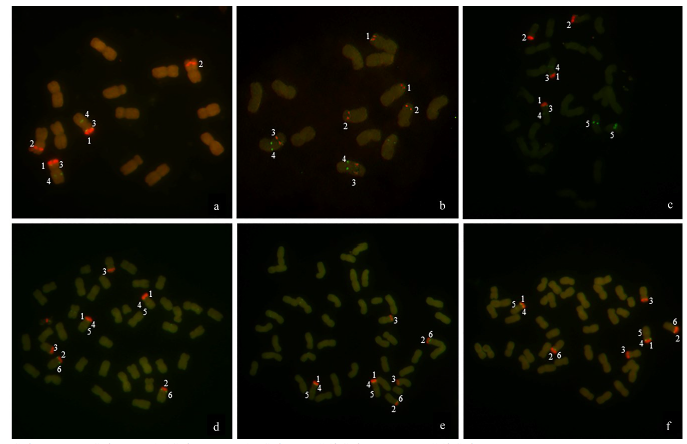
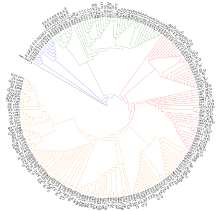
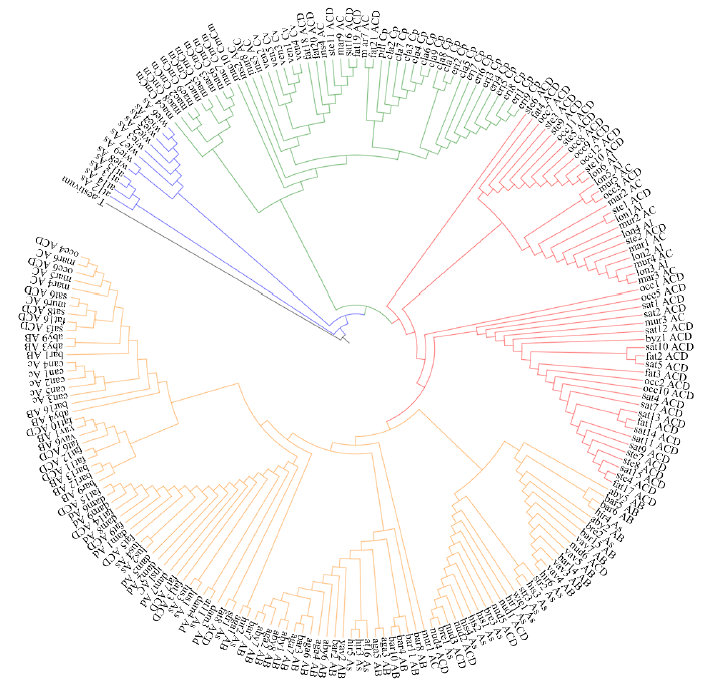
Chromosomal Localization of Ribosomal DNA and Phylogenetic Analysis of 45S rDNA in Avena
Wang Bingce1( ), Liu Xiaojuan1(
), Liu Xiaojuan1( ), Cheng Bin2, Ren Mingjian1, Xu Ruhong1, Zhang Suqin1, Zhang Liyi2, He Fang1(
), Cheng Bin2, Ren Mingjian1, Xu Ruhong1, Zhang Suqin1, Zhang Liyi2, He Fang1( )
)
- 1College of Agriculture, Guizhou University/Guizhou Subcenter of National Wheat Improvement Center, Guiyang 550025, Guizhou, China
2Institute of Upland Crops, Guizhou Academy of Agriculture Sciences, Guiyang 550006, Guizhou, China
| [1] | 李威, 周青平. 六种裸燕麦品种种子萌发期抗旱性的研究. 草业与畜牧, 2008,5(9):5-8. |
| [2] | 彭远英, 颜红海, 郭来春, 等. 燕麦属不同倍性种质资源抗旱性状评价及筛选. 生态学报, 2011,31(9):134-147. |
| [3] | 李英丽, 方正, 毛明艳. 不同燕麦品种耐碱性筛选和鉴定. 河北农业大学学报, 2014,37(6):13-17. |
| [4] | 董玉琛, 刘旭总. 中国作物及其野生近缘植物:粮食作物卷. 北京: 中国农业出版社, 2006. |
| [5] | Drossou A, Katsiotis A, Leggett J M, et al. Genome and species relationships in genus Avena based on RAPD and AFLP molecular markers. Theoretical and Applied Genetics, 2004,109(1):48-54. |
| [6] | Badaeva E D, Loskutov I G, Shelukhina O Y, et al. Cytogenetic analysis of diploid Avena L. species containing the as genome. Russian Journal of Genetics, 2005,41(12):1428-1433. |
| [7] | Rodionov A V, Tyupa N B, Kim E S, et al. Genomic configuration of the autotetraploid oat species avena macrostachya inferred from comparative analysis of ITS1 and ITS2 sequences:on the oat karyotype evolution during the early events of the Avena species divergence. Russian Journal of Genetics, 2005,41(5):518-528. |
| [8] | Loskutov I G. On evolutionary pathways of Avena species. Genetic Resources and Crop Evolution, 2008,55(2):211-220. |
| [9] | Nikoloudakis N, Skaracis G, Katsiotis A. Evolutionary insights inferred by molecular analysis of the ITS1-5.8S-ITS2 and IGS Avena sp. sequences. Molecular Phylogenetics and Evolution, 2008,46(1):102-115. |
| [10] | 刘青, 刘欢, 林磊. 燕麦属系统学研究进展. 热带亚热带植物学报, 2014(5):516-524. |
| [11] | 龚志云, 吴信淦, 程祝宽, 等. 水稻45S rDNA和5S rDNA的染色体定位研究. 遗传学报, 2002,29(3):241-244. |
| [12] | Pontes O, Cotrim H, Pais S, et al. Physical mapping,expression patterns and interphase organisation of rDNA loci in Portuguese endemic Silene cintrana and Silene rothmaleri. Chromosome Research, 2000,8(4):313-317. |
| [13] | Kato A, Lamb J C, Birchler J A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proceedings of the National Academy of Sciences, 2004,101(37):13554-13559. |
| [14] | Mahelka V, Kopecký D, Baum B R. Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Molecular Biology and Evolution, 2013,30(9):2065-2086. |
| [15] | Clegg M T, Zurawski G. Chloroplast DNA and the study of plant phylogeny:present status and future prospects//Soltis P S,Soltis D E,Doyle J J. Molecular Systematics of Plants. Springer:Boston,MA, 1992. |
| [16] | Peng Y Y, Wei Y M, Baum B R, et al. Molecular diversity of the 5S rRNA gene and genomic relationships in the genus Avena (Poaceae:Aveneae). Genome, 2008,51(2):137-154. |
| [17] | Yan H H, Baum B R, Zhou P P, et al. Phylogenetic analysis of the genus Avena based on chloroplast intergenic spacer psb A-trn H and single-copy nuclear gene Acc1. Genome, 2014,57(5):267-277. |
| [18] | Fu Y B. Oat evolution revealed in the maternal lineages of 25 Avena species. Scientific Reports, 2018,8(1):1-12. |
| [19] | Thompson J D, Gibson T J, Plewniak F, et al. The CLUSTAL_X Windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 1997,25(24):4876-4882. |
| [20] | Kumar S, Stecher G, Li M, et al. MEGA X:Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 2018,35(6):1547-1549. |
| [21] | Linc G, Gaál E, Molnár I, et al. Molecular cytogenetic (FISH) and genome analysis of diploid wheatgrasses and their phylogenetic relationship. PLoS ONE, 2017,12(3):e0173623. |
| [22] | Mahelka V, Kopecký D, Baum B R. Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Molecular Biology and Evolution, 2013,30(9):2065-2086. |
| [23] | 赵筱芳. 冰草居群45S rDNA基因位点FISH分析. 成都:四川农业大学, 2016. |
| [24] | Choi H W, Koo D H, Bang K H, et al. FISH and GISH analysis of the genomic relationships among Panax species. Genes and Genomics, 2009,31(1):99-105. |
| [25] | 符文炎, 刘义飞, 黄宏文. 荧光原位杂交技术在植物多倍体起源与进化研究中的应用. 热带亚热带植物学报, 2014,22(3):314-322. |
| [26] | Manns U, Anderberg A A. Molecular phylogeny of Anagallis (Myrsinaceae) based on ITS, trn L-F,and ndh F sequence data. International Journal of Plant Sciences, 2005,166(6):1019-1028. |
| [27] | Gillespie L J, Soreng R J, Bull R D, et al. Phylogenetic relationships in subtribe Poinae (Poaceae,Poeae) based on nuclear ITS and plastid trn T-trn L-trn F sequences. Botany, 2008,86(8):938-967. |
| [28] | Attar F, Riahi M, Daemi F, et al. Preliminary molecular phylogeny of Eurasian Scrophularia (Scrophulariaceae) based on DNA sequence data from trnS-trnG and ITS regions. Plant Biosystems, 2011,145(4):857-865. |
| [1] | Zhu Xu, Hu Weili, Yang Houyong, Xu Yang, Xiang Zhen, Yang Ling, Yang Pengcheng. Analysis of Suitable Agronomic Traits for Mechanized Harvesting Mung Bean Varieties (Lines) in Nanyang Basin [J]. Crops, 2021, 37(4): 93-98. |
| [2] | Qu Xiangchun, Wang Nai, Shi Guishan, Yu Miao, Li Haiqing, Gao Yue, Xu Ning, Chen Bingru. Application in Similarity-Difference Analysis Method on Evaluation of Grain Sorghum Hybrids [J]. Crops, 2021, 37(3): 46-50. |
| [3] | Zhou Yuexia, Fan Yu, Ruan Jingjun, Yan Jun, Lai Dili, Peng Yan, Tang Yong, Weng Wenfeng, Cheng Jianping. Correlation Analysis of Oat Grain Nutrition and Agronomic Traits [J]. Crops, 2021, 37(2): 165-172. |
| [4] | Jin Jiangang, Tian Zaifang. Grey Correlation Analysis of Introduced Tartary Buckwheat in the Northern Shanxi [J]. Crops, 2021, 37(2): 52-56. |
| [5] | Wang Yujiao, Cao Qi, Chang Xuhong, Wang Demei, Wang Yanjie, Yang Yushuang, Zhao Guangcai, Shi Shubing. Effects of Chemical Regulation on Wheat Yield and Quality under Different Soil Conditions [J]. Crops, 2021, 37(2): 96-100. |
| [6] | Duan Huimin, Lu Xiao, Zhou Xiaojie, Li Gaofeng, Wen Guohong, Wang Yuping, Cheng Lixiang, Zhang Feng. Effects of Potato Leaf Type and Planting Density on Yield Components [J]. Crops, 2021, 37(1): 160-167. |
| [7] | Pan Xiaoxue, Hu Mingyu, Wang Zhongwei, Wu Hong, Lei Kairong. Evaluation of Agronomic Traits and Cold Tolerance at Germination Stage in Rice (Oryza sativa L.) Germplasms [J]. Crops, 2021, 37(1): 47-53. |
| [8] | Yang Wanjun, Pan Xiangyu, Wang Xiuhua, Wang Lu, Zhao Yan. Genetic Diversity Analysis of Yield and Agronomic Traits of 119 Alfalfa Varieties (Lines) [J]. Crops, 2020, 36(6): 17-22. |
| [9] | Yang Xuele, Zhang Lu, Li Zhiqing, He Luqiu. Diversity Analysis of Tartary Buckwheat Germplasms Based on Phenotypic Traits [J]. Crops, 2020, 36(5): 53-58. |
| [10] | Chen Weiguo, Zhang Zheng, Shi Yugang, Cao Yaping, Wang Shuguang, Li Hong, Sun Daizhen. Drought-Tolerance Evaluation of 211 Wheat Germplasm Resources [J]. Crops, 2020, 36(4): 53-63. |
| [11] | Zhao Yanhong,Hou Wenhuan,Liao Xiaofang,Tang Xingfu,Li Chuying. Effects of Different Sunshine Durations on Main Agronomic Traits of Roselle [J]. Crops, 2020, 36(2): 172-175. |
| [12] | Chen Tianxin,Wang Yanjie,Zhang Yan,Chang Xuhong,Tao Zhiqiang,Wang Demei,Yang Yushuang,Zhu Yingjie,Liu Akang,Shi Shubing,Zhao Guangcai. Effects of Different Nitrogen Rates on Photosyntheticand Physiological Indexes and Yield of Winter Wheat [J]. Crops, 2020, 36(2): 88-96. |
| [13] | Wang Heying,Guo Xiaohong,Zhang Qinming,Ma Yan,Li Meng,Jiang Hongfang,Hu Yue,Lan Yuchen,Xu Lingqi,Guo Hongtao,Lü Yandong. Effects of Sowing in Line under Water on Agronomic Characters and Yield Components of Rice in Cold Region [J]. Crops, 2020, 36(1): 81-88. |
| [14] | Cao Tingjie,Zhang Yu’e,Hu Weiguo,Yang Jian,Zhao Hong,Wang Xicheng,Zhou Yanjie,Zhao Qunyou,Li Huiqun. Detection of Three Dwarfing Genes in the New Wheat Cultivars (Lines) Developed in South Huang-Huai Valley and Its Association with Agronomic Traits [J]. Crops, 2019, 35(6): 14-19. |
| [15] | Zhang Ting,Lu Lahu,Yang Bin,Yuan Kai,Zhang Wei,Shi Xiaofang. Comparative Analysis of Wheat Agronomic Traits in Four Provinces of Huanghuai Wheat Area [J]. Crops, 2019, 35(6): 20-26. |
|
||