植物在生长过程中经常受到非生物胁迫,如干旱、热、冷、营养缺乏和土壤中过量盐或有毒金属,这些非生物胁迫限制了世界范围内耕地的利用,并对作物生产力产生了负面影响[1]。土壤盐渍化是现代农业面临的主要问题之一,不仅会引起离子毒性和渗透胁迫,还会导致植物缺乏养分,严重影响植物生长发育,进而影响品质及产量。与其他仅影响作物某一特定时期的胁迫不同,盐胁迫会贯穿作物的整个生命周期[2]。因此,了解植物如何感知盐胁迫信号并适应不利环境条件至关重要。在植物长期进化过程中,已形成一套响应盐胁迫的调控机制。当受到盐胁迫时,植物首先在基因转录水平上进行调控,然后通过控制代谢合成进行调节,应对逆境带来的损伤。随着高通量测序技术的快速发展,转录组测序已成为研究基因差异表达的重要手段[3],对在转录水平揭示植物响应盐胁迫的调控机制具有重要意义。
研究表明,bHLH转录因子在响应盐胁迫方面发挥重要功能。在盐胁迫下,甜菜BvbHLH93基因在根和叶中的表达量显著上调[6];高盐下,水稻OsbHLH148的转录水平会迅速提高[7]。除此之外,bHLH转录因子是植物盐胁迫的正调控因子。对转基因烟草进行表型分析发现,过表达NtbHLH123基因可增强烟草对盐胁迫的抗性,而NtbHLH123沉默则植株对盐胁迫的耐受性降低,表明NtbHLH123正调控烟草耐盐性[8];对ZmbHLH91过表达植株进行高盐处理,发现转基因植株具有更好的生长状态和绿叶率,另外,在胁迫处理后,转基因植株过氧化物酶(POD)具有更高的活性,说明ZmbHLH91在高盐以及渗透胁迫中可能是一个正向的胁迫响应因子,可能通过提高POD活性,清除植物体内过氧化物来提高抗逆性[9];在小麦中,过表达TabHLH39可提高植物对高盐处理的抗性[10]。盐胁迫下,葡萄VvbHLH1的过表达会导致类黄酮生物合成路径、脱落酸信号通路和胁迫反应等相关基因的上调表达,推测VvbHLH1可能通过调控黄酮类化合物含量来增强植株耐盐性[11];拟南芥AtbHLH112通过与E-box和GCG-box基序结合,调控非生物胁迫相关基因的表达,以增加脯氨酸含量、减少活性氧积累和水分流失来提高抗盐能力[12]。
本研究利用RNA-Seq方法对盐胁迫处理前后的科农199幼苗根系进行转录组测序,鉴定小麦根系响应盐胁迫应答基因,并分别对差异表达基因(differentially expressed gene,DEG)进行GO分析和KEGG注释和富集分析,从转录水平揭示小麦bHLH转录因子响应盐胁迫的分子机制,进一步筛选小麦bHLH转录因子耐盐相关基因,为解析小麦耐盐性状的分子机制奠定基础。
1 材料与方法
1.1 试验材料
以中国农业科学院作物科学研究所提供的冬小麦品种科农199为材料,其根系发达,尤其生育后期根系活力强,具有较强吸收深层土壤水分养分的能力,适应性强,抗逆能力强。采用水培法进行培养,培养条件为光照16 h、黑暗8 h、22 ℃下水培养4 d。用花无缺营养液培养至两叶一心时期后改用200 mmol/L的NaCl溶液处理。分别在0、1和6 h采集根组织样本,设3个重复,取样后立即于液氮中速冻,存于-80 ℃冰箱用于后续提取RNA[13],并对每个重复进行RNA-Seq分析。
1.2 小麦bHLH转录因子家族在染色体上的定位
从Pfam数据库(
1.3 小麦bHLH转录因子家族系统进化树的构建
从Wheat Omics1.0数据库(
1.4 小麦bHLH转录因子差异基因表达分析
在盐胁迫处理科农199得到的转录组数据中筛选到小麦489个bHLH转录因子,以|log2Fold- change|≥2且FDR<0.01为筛选标准,比对得到差异表达基因后进行分析。
1.5 差异表达基因的GO富集分析
将筛选后的小麦bHLH转录因子家族的差异表达基因提交到Agrigo(GO analysis Toolkit and Database for Agricultural Community)(
1.6 差异表达基因的KEGG通路分析
使用KOBAS网站(
1.7 差异表达基因的实时荧光定量PCR验证
为了验证转录组数据得到的DEG可靠性,分别选择盐处理1和6 h后都上调的4个DEG和都下调的4个DEG进行qRT-PCR验证,根据小麦数据库下载的序列,用网站中的PrimerServer (BETA): PCRPrimersBatch Design&SpecificityCheck工具设计荧光定量PCR特异引物(表1)。以盐胁迫处理后0、1、6 h的根部组织为样本,以处理0 h为对照,使用罗氏荧光定量PCR仪分析差异表达基因的相对表达水平。反应程序设置为95 ℃,10 s;95 ℃ 5 s,60 ℃ 30 s,72 ℃ 10 s,40个循环。每组试验设置3个生物学样本,每个生物学样本设置3个技术重复,数据为3个生物学重复平均值±标准误差。采用2-ΔΔCT方法计算相对表达量,选择TaGAPDH作为内参基因。采用SPSS软件进行数据分析。
表1 引物信息
Table 1
| 引物名称Primer name | 正向引物(5’-3’)Forward primer | 反向引物(3’-5’)Reverse primer |
|---|---|---|
| TraesCS5B01G518800 | GATCATGGTAGCCCGTCACC | TTCACCATCACGTTCCCCTC |
| TraesCS1A01G110400 | CACCTTGTTAGCTTGTGTGGTG | GCAATGGACGGTGATCTCGTTA |
| TraesCS4A01G408800 | GAGGCAAAGCTCTCGGAGG | TCTTCCACCTTTGCCATGGT |
| TraesCS5D01G244000 | CCTTTCCTTCCGTTTCTGTCG | ATGCGGTAACAACGACGACA |
| TraesCS2B01G543800 | TCGACTTCCTGCACCTTTGG | CTGCAGATATAGTCCCGCCG |
| TraesCS1A01G345200 | GCAATCTGTGACGAGTCGGA | TACCGCTCTTCCTTGCAGTC |
| TraesCS7B01G074900 | GGATATGACGAACCAAGAACAGC | CATCGATGGAGTAACAGCACTG |
| TraesCS1B01G359000 | TGCATCTCGTGATCTCCAAGT | TAACCACTCTTCCTAGCGGC |
| TraesCS6D02G196300 (TaGAPDH) | TTAGACTTGCGAAGCCAGCA | AAATGCCCTTGAGGTTTCCC |
1.8 共同差异表达基因的表达聚类分析
从2组差异表达基因中筛选出公共的差异表达基因,利用R软件的Pheatmap包(
2 结果与分析
2.1 小麦bHLH家族转录因子染色体分布
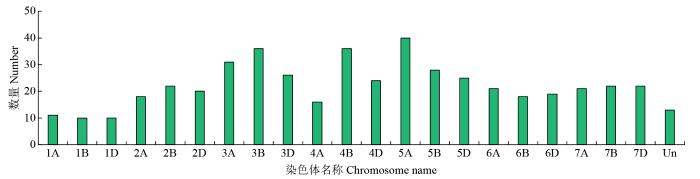
通过对bHLH保守结构域进行比对分析,在小麦基因组上共鉴定到489个bHLH转录因子。染色体定位分析结果(图1)显示,这些基因分布在21条染色体上,其中3A、3B、4B以及5A染色体上的bHLH转录因子数目较多,而在1A、1B和1D染色体上的分布则相对较少。
图1
图1
小麦bHLH家族转录因子在染色体上分布
Fig.1
The distribution of the wheat bHLH family transcription factors in the chromosomes
2.2 小麦bHLH家族转录因子进化树分析
2.3 基因表达差异分析
为了挖掘响应盐胁迫的小麦bHLH转录因子基因,对盐处理1 h及6 h后的DEG分析(表2)表明,小麦bHLH转录因子在盐处理1 h后的DEG数目为100,其中63个基因表现为上调趋势,37个下调表达;而在盐处理6 h后有148个DEG,其中91个基因上调表达,57个下调表达;经筛选后发现,共有44个DEG同时存在于盐胁迫1 h和6 h处理后,其中17个基因上调表达,27个下调表达。
表2 44个共同差异表达基因的调控模式
Table 2
| 基因号 Gene ID | 染色体位置 Chromosome location | 调控模式 Regulation pattern | 基因号 Gene ID | 染色体位置 Chromosome location | 调控模式 Regulation pattern |
|---|---|---|---|---|---|
| TraesCS4A01G408800 | 681653116~681658841 | 上调 | TraesCS4B01G308300 | 599168078~599170653 | 上调 |
| TraesCS4B01G308200 | 598910941~598914019 | 上调 | TraesCS5A01G237500 | 453519135~453519842 | 上调 |
| TraesCS5D01G244000 | 352453691~352454917 | 上调 | TraesCS5A01G401300 | 594132650~594134217 | 上调 |
| TraesCS2D01G270300 | 334040936~334043454 | 上调 | TraesCS4D01G306400 | 474700560~474701269 | 上调 |
| TraesCS5B01G518800 | 681737539~681740186 | 上调 | TraesCS1A01G110400 | 110072873~110073869 | 上调 |
| TraesCS2B01G289900 | 402217632~402220229 | 上调 | TraesCS4B01G304500 | 592819415~592821925 | 下调 |
| TraesCS7B01G211600 | 387160478~387163988 | 上调 | TraesCS2B01G463800 | 657790256~657793959 | 下调 |
| TraesCS3B01G002700 | 2169304~2172368 | 上调 | TraesCS7A01G229900 | 200230140~200231461 | 下调 |
| TraesCS5B01G235200 | 415127011~415128223 | 上调 | TraesCS6B01G244800 | 437162916~437168918 | 下调 |
| TraesCS5D01G411600 | 475000589~475001987 | 上调 | TraesCS4A01G292800 | 595294248~595297051 | 下调 |
| TraesCS4D01G306300 | 474598568~474603146 | 上调 | TraesCS4A01G404700 | 678217690~678220066 | 下调 |
| TraesCS2A01G271700 | 444623265~444626004 | 上调 | TraesCS4D01G328800 | 487119229~487120910 | 下调 |
| TraesCS6D01G197500 | 275152959~275163740 | 下调 | TraesCS2D01G517000 | 607986836~607997092 | 下调 |
| TraesCS4D01G302700 | 470506568~470509154 | 下调 | TraesCS2B01G543800 | 741241659~741248415 | 下调 |
| TraesCS6A01G190600 | 254936294~254937809 | 下调 | TraesCS1A01G345200 | 532672290~532673440 | 下调 |
| TraesCS6A01G214900 | 394613029~394618337 | 下调 | TraesCS2B01G298600 | 418276647~418278407 | 下调 |
| TraesCS3B01G550000 | 785039358~785043172 | 下调 | TraesCS2A01G281200 | 469810800~469812561 | 下调 |
| TraesCS4D01G018700 | 7992189~7995548 | 下调 | TraesCS2D01G280100 | 351981513~351983269 | 下调 |
| TraesCS3D01G495700 | 587745410~587748716 | 下调 | TraesCS5A01G555300 | 706705261~706706358 | 下调 |
| TraesCS3B01G550200 | 785280464~785284379 | 下调 | TraesCS3A01G489600 | 717006915~717011250 | 下调 |
| TraesCS3A01G489700 | 717296946~717300622 | 下调 | TraesCS1B01G359000 | 588448000~588449096 | 下调 |
| TraesCS3D01G495600 | 587644739~587648644 | 下调 | TraesCS7B01G074900 | 84388710~84389462 | 下调 |
2.4 差异表达基因的GO富集分析
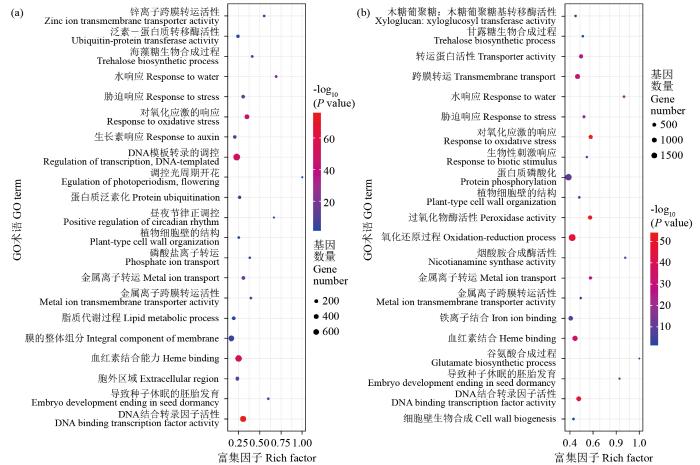
分别对盐胁迫处理后0、1和6 h的差异表达基因进行GO富集分析。根据GO功能分类,这些差异表达基因可分为分子功能、生物过程和细胞组成3个方面的亚类。结果显示,与处理后0 h对照相比,处理1 h的DEGs主要富集在DNA结合转录因子活性、抗氧化反应、转录调控及DNA模块化、调控光周期和昼夜节律等分子功能方面;另外在胁迫应答、水盐响应、生长素响应、蛋白质泛素化、磷酸离子运输及金属离子运输和脂质代谢等生物过程中也有部分DEGs富集;细胞组成类主要分布在植物细胞壁组织、膜整体组分和胞外区等分支(图2a)。而处理6 h的DEGs主要注释在跨膜转运、蛋白磷酸化、细胞壁生物起源、氧化还原及铁离子结合等生物过程中(图2b)。
图2
图2
差异表达基因的GO分析
圆点位置代表代谢途径;圆点大小代表基因的数量;Rich factor为该代谢路径下差异基因数目与所有注释到该路径基因数目的比值,数值越大表示富集程度越大。下同。
Fig.2
GO analysis of differentially expressed genes
The dots represent metabolic pathways; The size of the dot represents the number of genes; Rich factor is the ratio of the number of differential genes in this metabolic pathway to the number of genes annotated to this pathway, and the higher values indicate a higher degree of enrichment. The same below.
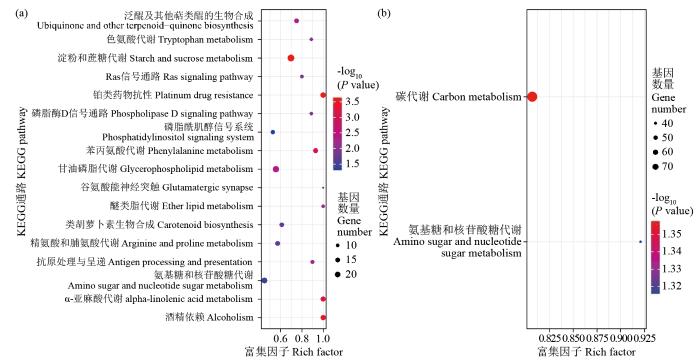
2.5 差异表达基因的KEGG通路分析
图3
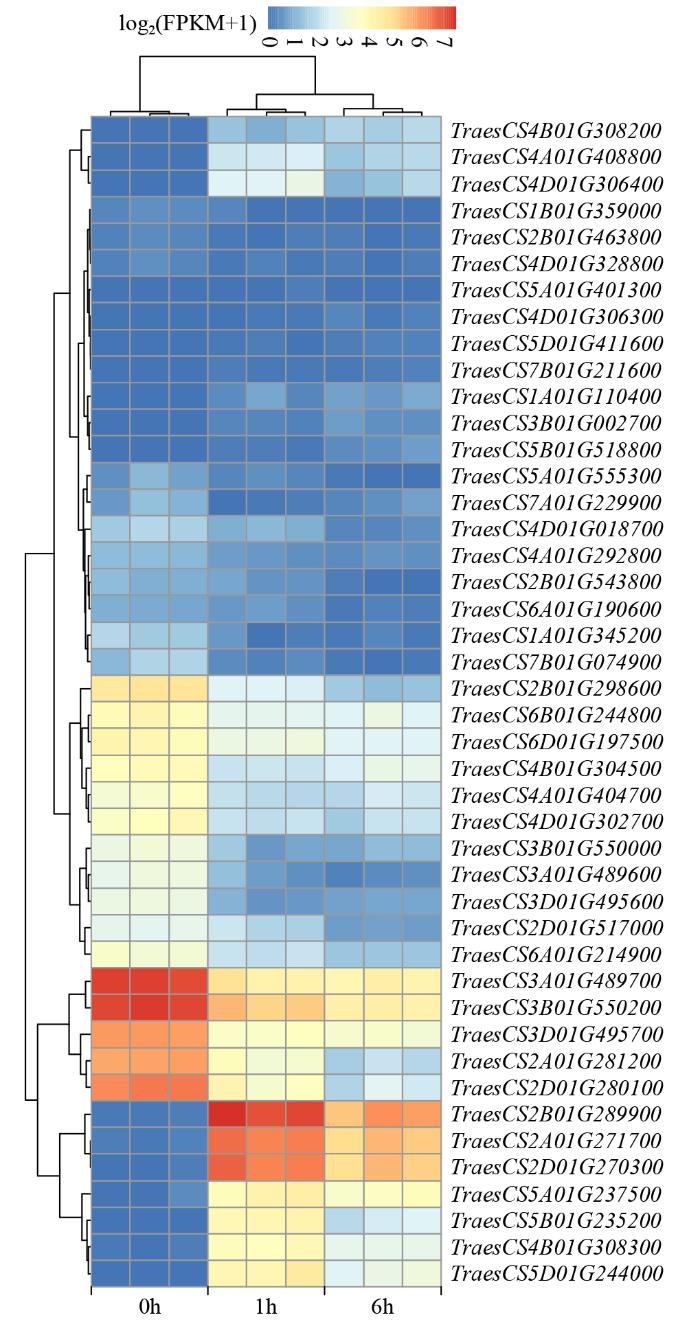
2.6 bHLH转录因子家族差异表达基因的表达聚类分析
为了进一步明确bHLH转录因子家族差异表达基因的表达变化,从2组差异表达基因中(盐处理1 h和6 h后)挑选公共的差异表达基因进行表达聚类分析。结果(图4)表明,小麦bHLH转录因子中有44个基因在盐胁迫下表达量发生变化,相比盐处理0 h来说,对盐处理1和6 h后其表达量有明显上升或下降的趋势,其中上调表达的基因有17个;而下调表达的基因有27个。
图4
图4
小麦响应盐胁迫bHLH转录因子聚类分析
Fig.4
Cluster analysis of wheat bHLH transcription factor genes in response to salt stress
2.7 差异表达基因的实时荧光定量PCR验证
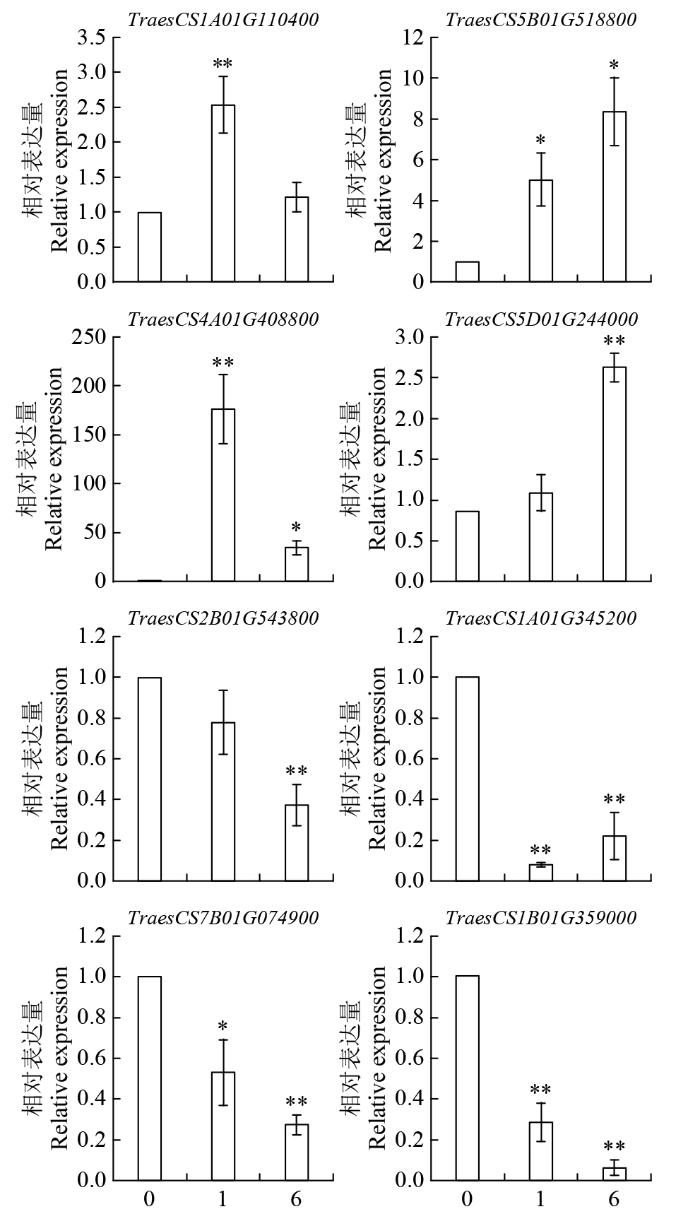
为了验证转录组数据结果的可靠性,随机选取8个差异表达基因进行qRT-PCR分析。由图5可看出,8个基因在胁迫处理后表达发生明显改变,上调表达的4个基因中,TraesCS1A01G110400和TraesCS4A01G408800在盐处理1 h时极显著上调,处理6 h后表达上调趋势较处理1 h来说相对低;而TraesCS5B01G518800和TraesCS5D01G244000的表达量则是随着盐胁迫处理时间延长逐渐上升,这与转录组测序数据中差异基因的表达量趋势一致。另外4个下调表达的基因,其表达变化趋势也与转录组数据结果一致,TraesCS2B01G543800、TraesCS7B01G074900、TraesCS1B01G359000的表达量都随盐胁迫处理时间逐渐下降,TraesCS1A01G345200的表达量在盐胁迫处理1 h时显著下降,当盐处理6 h时其表达量变化趋势反而没有1 h盐处理的表达量下降趋势明显。虽然qRT-PCR分析得到的表达差异与转录组测序分析的表达差异倍数不一致,但胁迫诱导表达的变化趋势相同,在一定程度上说明了转录组测序结果的可靠性。
图5
图5
部分DEGs的实时荧光定量PCR验证
“*”:差异显著(0.05 > P > 0.01);“**”:差异极显著(P < 0.01)。
Fig.5
Real-time PCR validation of part of DEG
“*”: the difference is significant (0.05 > P > 0.01);“**”: the difference is significant (P < 0.01).
3 讨论
小麦是我国重要的粮食作物,在生长发育过程中会遭受各种非生物胁迫,严重影响其产量与品质。土壤盐碱化严重制约植物的生长进程,盐碱地是我国主要的后备土壤资源,约80%的盐渍土地有待开发利用。因此,研究植物响应盐胁迫过程,培育具有一定耐盐性的作物品种,结合盐碱地的开发利用,可在一定程度上增加耕地面积,保障粮食安全。
研究[16]表明,bHLH转录因子通过影响植物的离子平衡、膜透性来调节植物对盐胁迫的耐受性。对AhbHLH18进行盐胁迫处理后,其表达发生明显变化,在12 h内,AhbHLH18的相对表达量处于上升阶段,12 h后逐渐趋于稳定,并且相比于0 h,6 h的相对表达量表现为显著差异,12~48 h的表达量为极显著差异。对盐胁迫条件下的粗山羊草进行转录组测序,得到546个和876个上调和下调表达的DEG,通过GO和KEGG富集分析和注释,明确了响应盐胁迫DEG的生物功能和所富集的信号通路[17]。以甜菜耐盐品系T710MU为研究对象,通过对转录组测序数据进行分析获得10个响应盐胁迫的bHLH基因家族的差异表达基因,荧光定量PCR结果显示,在盐处理12 h后,叶片中表达量上调,推测这些基因为盐应答转录因子[18]。对毛竹bHLH转录因子进行转录组表达谱分析,结果表明有151个PebHLHs在不同组织和生长发育时期有不同程度表达,表明其在毛竹中功能的多样性,盐胁迫处理后,毛竹叶片中同一基因表达的差异以及不同PebHLHs基因间表达量的差异进一步说明PebHLHs功能的多样性和复杂性[19]。以上结论在一定程度上表明,不同物种在响应盐胁迫时,基因在表达量上表现为多样性和复杂性,进一步发挥不同功能。
bHLH家族作为植物体内第二大转录因子,普遍存在于大多数真核生物中,不仅在植物生长发育各个阶段、下胚轴伸长、种子萌发、气孔开闭及开花等方面发挥重要功能,还在昼夜节律、生物合成、信号传导及非生物胁迫中发挥功能。目前,已经在白菜[20]、甘薯[21]、谷子[22]、西瓜[23]及拟南芥等植物中鉴定了bHLH转录因子家族,通过生物信息学方法分析bHLH基因的结构和蛋白性质,并对其功能进行预测。但在小麦中,还没有对bHLH转录因子家族进行鉴定,本研究利用生物信息学分析的方法,鉴定出489个TabHLH基因,染色体定位发现其分布在21条染色体上,对盐胁迫下TabHLH基因的转录组分析发现,在盐处理1和6 h后分别有100个和148个DEGs参与盐胁迫应答反应。差异表达基因的GO富集分析结果显示,富集程度最高的GO功能分类项为DNA结合转录因子活性以及转录调控等分子功能,这从一定程度上证明了植物分子功能是造成植物响应盐胁迫差异性的主要原因之一。KEGG注释分析将差异表达基因富集在淀粉和蔗糖代谢途径以及氨基糖和核苷酸糖代谢途径上,这些途径可在植物生长发育过程中提供能量。表达聚类分析表明,盐胁迫下小麦bHLH家族转录因子具有特异性表达,可为挖掘耐盐基因提供依据。
4 结论
通过转录组测序手段和生物信息学分析的方法对小麦bHLH转录因子在盐胁迫下的表达特性进行了系统分析。共鉴定出489个bHLH转录因子,分布在21条染色体上;盐胁迫下小麦根部组织转录组测序结果表明,在盐胁迫处理后1和6 h差异表达基因(DEG)共有44个,其中上调表达基因17个,下调表达有27个;GO分析表明,DEG富集在水盐胁迫及生长素调节等方面;KEGG注释分析表明,DEG主要富集在淀粉和蔗糖代谢途径、氨基糖和核苷酸糖代谢等信号通路。
参考文献
Abiotic stress responses in plants
DOI:10.1038/s41576-021-00413-0 [本文引用: 1]
Adaptive strategy of allohexaploid wheat to long-term salinity stress
DOI:10.1186/s12870-020-02423-2
PMID:32397960
[本文引用: 1]

Most studies of crop salinity tolerance are conducted under short-term stress condition within one growth stage. Understanding of the mechanisms of crop response to long-term salinity stress (LSS) is valuable for achieving the improvement of crop salinity tolerance. In the current study, we exposed allohexaploid wheat seeds to LSS conditions from germination stage to young seedling stage for 30 days. To elucidate the adaptive strategy of allohexaploid wheat to LSS, we analyzed chloroplast ultrastructure, leaf anatomy, transcriptomic profiling and concentrations of plant hormones and organic compatible solutes, comparing stressed and control plants.Transcriptomic profiling and biochemical analysis showed that energy partitioning between general metabolism maintenance and stress response may be crucial for survival of allohexaploid wheat under LSS. Under LSS, wheat appeared to shift energy from general maintenance to stress response through stimulating the abscisic acid (ABA) pathway and suppressing gibberellin and jasmonic acid pathways in the leaf. We further distinguished the expression status of the A, B, and D homeologs of any gene triad, and also surveyed the effects of LSS on homeolog expression bias for salinity-tolerant triads. We found that LSS had similar effects on expression of the three homeologs for most salinity-tolerant triads. However, in some of these triads, LSS induced different effects on the expression of the three homeologs.The shift of the energy from general maintenance to stress response may be important for wheat LSS tolerance. LSS influences homeolog expression bias of salinity-tolerant triads.
NaCl胁迫下黑果枸杞转录组测序分析
DOI:10.13560/j.cnki.biotech.bull.1985.2019-0492
[本文引用: 1]

研究黑果枸杞在不同浓度盐胁迫下基因表达谱变化情况,为进一步研究黑果枸杞抗盐分子机制奠定研究基础。对0(CK)、50、250 mmol/L NaCl溶液胁迫的黑果枸杞组培苗的根和叶在胁迫时间为0、1、12 h时分别取样,采用转录组测序(RNA-Seq)技术进行测序分析。结果表明,转录组测序共产生 222.49 Gb 原始数据,拼接出 Unigenes 86 037条,注释到 7大功能数据库(GO、KEGG、KOG、NR、Pfam、Swiss-Prot和egg NOG)上的 Unigenes 总数为46 594个,占总Unigenes 的54.76%,还有38 929个Unigenes在这些数据库中没有得到注释。通过 GO 分类和 KEGG Pathway 富集性分析,分别归于 51 个 GO 类别和 211条代谢途径。差异表达基因分析显示,黑果枸杞叶片和根的上调基因和下调基因数随着NaCl浓度的增大和处理时间的延长均呈增加趋势,叶片中的上调基因数(7 514)小于下调基因数(9 032),根中的上调基因数(12 347)大于下调基因数(11 559)。在黑果枸杞盐胁迫下转录组中发现28 325个SSR位点,最多的为单核苷酸 SSR,占 70.47%。综合分析表明,黑果枸杞对盐胁迫的反应是一个多基因参与、多个生物过程协同调控的过程,基因表达量的变化可能是基因调控的主要方式。
Basic helix-loop-helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis
DOI:10.3390/ijms22137152
URL
[本文引用: 1]

The basic helix-loop-helix (bHLH) transcription factor family is one of the largest transcription factor gene families in Arabidopsis thaliana, and contains a bHLH motif that is highly conserved throughout eukaryotic organisms. Members of this family have two conserved motifs, a basic DNA binding region and a helix-loop-helix (HLH) region. These proteins containing bHLH domain usually act as homo- or heterodimers to regulate the expression of their target genes, which are involved in many physiological processes and have a broad range of functions in biosynthesis, metabolism and transduction of plant hormones. Although there are a number of articles on different aspects to provide detailed information on this family in plants, an overall summary is not available. In this review, we summarize various aspects of related studies that provide an overview of insights into the pleiotropic regulatory roles of these transcription factors in plant growth and development, stress response, biochemical functions and the web of signaling networks. We then provide an overview of the functional profile of the bHLH family and the regulatory mechanisms of other proteins.
Sugar beet BvBHLH 92 tissue expression and subcellular localization
玉米转录因子ZmbHLH91对非生物逆境胁迫的应答
DOI:10.3724/SP.J.1006.2022.13060
[本文引用: 1]

bHLH (basic helix-loop-helix)是植物中一个重要的转录因子家族, 在调控植物生长发育、逆境胁迫及信号转导过程中发挥着重要作用。目前, 动物中大部分bHLH转录因子功能已明确, 但是在植物中, 尤其是玉米中的研究报道较少。在前期工作中, 我们对玉米生长发育过程中的4个关键时期进行了根系表型鉴定和转录组测序分析, 发现转录因子ZmbHLH91在六叶期(V6)、十二叶期(V12)和抽雄期(VT)的相邻时期间的表达均差异显著, 且ZmbHLH91在V6, V12和VT这些根系生长发育活跃期的表达量较高, 推测该基因可能调控玉米根系的生长发育。为探究ZmbHLH91基因在根系生长和抵抗逆境胁迫方面的作用, 本研究克隆了ZmbHLH91 (AC: NC_AQL05369)基因, 该基因全长2112 bp, 具有bHLH转录因子家族特有的保守结构域。实时荧光定量PCR (RT-qPCR)分析表明, ZmbHLH91在玉米根中的表达量最高, 其在三叶期幼根中的表达量高于抽雄期成熟根。在高盐、渗透、低温以及脱水胁迫处理下, 玉米幼苗中ZmbHLH91的表达均上调。在无胁迫处理的1/2 MS培养基上, ZmbHLH91异源表达拟南芥与野生型拟南芥的根长无明显差别, 而在梯度浓度NaCl和甘露醇处理的培养基上, ZmbHLH91异源表达拟南芥的根均长于野生型, 且差异显著; 在土壤中进行干旱和高盐处理后, 转基因拟南芥比野生型拟南芥表现出更好的生长状态、更高的过氧化物酶(POD)活性和更高的绿叶率。推测ZmbHLH91基因可能参与响应高盐、干旱以及渗透胁迫等逆境条件。ZmbHLH91基因在茉莉酸(jasmonic acid, JA)、脱落酸(abscisic acid, ABA)等激素处理下均上调表达, 推测ZmbHLH91基因可能参与响应JA和ABA激素信号。在梯度浓度JA处理的培养基上, 转基因拟南芥的根长均长于野生型, 且差异显著。酵母双杂交实验表明ZmbHLH91与ZmMYC2相互作用, ZmMYC2是JA信号通路中重要的蛋白, 由此推测ZmbHLH91蛋白可能参与JA信号通路。综上所述, ZmbHLH91可能参与高盐、干旱和渗透胁迫应答及JA信号转导途径。本研究为进一步解析ZmbHLH91在玉米中的生物学功能提供了重要的参考依据。
The wheat transcription factor, TabHLH39, improves tolerance to multiple abiotic stressors in transgenic plants
DOI:S0006-291X(16)30577-0
PMID:27091431
[本文引用: 1]

Although bHLH transcription factors play important roles regulating plant development and abiotic stress response and tolerance, few functional studies have been performed in wheat. In this study, we isolated and characterized a bHLH gene, TabHLH39, from wheat. The TabHLH39 gene is located on wheat chromosome 5DL, and the protein localized to the nucleus and activated transcription. TabHLH39 showed variable expression in roots, stems, leaves, glumes, pistils and stamens and was induced by polyethylene glycol, salt and cold treatments. Further analysis revealed that TabHLH39 overexpression in Arabidopsis significantly enhanced tolerance to drought, salt and freezing stress during the seedling stage, which was also demonstrated by enhanced abiotic stress-response gene expression and changes to several physiological indices. Therefore, TabHLH39 has potential in transgenic breeding applications to improve abiotic stress tolerance in crops.Copyright © 2016 Elsevier Inc. All rights reserved.
The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis
DOI:10.1007/s00425-016-2489-3 URL [本文引用: 1]
Transcriptional regulation of bHLH during plant response to stress
DOI:S0006-291X(18)31625-5
PMID:30057319
[本文引用: 1]

Basic helix-loop-helix protein (bHLH) is the most extensive class of transcription factors in eukaryotes, which can regulate gene expression through interaction with specific motif in target genes. bHLH transcription factor is not only universally involved in plant growth and metabolism, including photomorphogenesis, light signal transduction and secondary metabolism, but also plays an important role in plant response to stress. In this review, we discuss the role of bHLH in plants in response to stresses such as drought, salt and cold stress. To provide a strong evidence for the important role of bHLH in plant stress response, in order to provide new ideas and targets for the prevention and treatment of plant stress resistance.Copyright © 2018 Elsevier Inc. All rights reserved.
The Arabidopsis basic/ helix-loop-helix transcription factor family
DOI:10.1105/tpc.013839
URL
[本文引用: 1]

The basic/helix-loop-helix (bHLH) proteins are a superfamily of transcription factors that bind as dimers to specific DNA target sites and that have been well characterized in nonplant eukaryotes as important regulatory components in diverse biological processes. Based on evidence that the bHLH protein PIF3 is a direct phytochrome reaction partner in the photoreceptor's signaling network, we have undertaken a comprehensive computational analysis of the Arabidopsis genome sequence databases to define the scope and features of the bHLH family. Using a set of criteria derived from a previously defined consensus motif, we identified 147 bHLH protein–encoding genes, making this one of the largest transcription factor families in Arabidopsis. Phylogenetic analysis of the bHLH domain sequences permits classification of these genes into 21 subfamilies. The evolutionary and potential functional relationships implied by this analysis are supported by other criteria, including the chromosomal distribution of these genes relative to duplicated genome segments, the conservation of variant exon/intron structural patterns, and the predicted DNA binding activities within subfamilies. Considerable diversity in DNA binding site specificity among family members is predicted, and marked divergence in protein sequence outside of the conserved bHLH domain is observed. Together with the established propensity of bHLH factors to engage in varying degrees of homodimerization and heterodimerization, these observations suggest that the Arabidopsis bHLH proteins have the potential to participate in an extensive set of combinatorial interactions, endowing them with the capacity to be involved in the regulation of a multiplicity of transcriptional programs. We provide evidence from yeast two-hybrid and in vitro binding assays that two related phytochrome-interacting members in the Arabidopsis family, PIF3 and PIF4, can form both homodimers and heterodimers and that all three dimeric configurations can bind specifically to the G-box DNA sequence motif CACGTG. These data are consistent, in principle, with the operation of this combinatorial mechanism in Arabidopsis.
Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae
DOI:10.1104/pp.110.153593
URL
[本文引用: 1]

Basic helix-loop-helix proteins (bHLHs) are found throughout the three eukaryotic kingdoms and constitute one of the largest families of transcription factors. A growing number of bHLH proteins have been functionally characterized in plants. However, some of these have not been previously classified. We present here an updated and comprehensive classification of the bHLHs encoded by the whole sequenced genomes of Arabidopsis (Arabidopsis thaliana), Populus trichocarpa, Oryza sativa, Physcomitrella patens, and five algae species. We define a plant bHLH consensus motif, which allowed the identification of novel highly diverged atypical bHLHs. Using yeast two-hybrid assays, we confirm that (1) a highly diverged bHLH has retained protein interaction activity and (2) the two most conserved positions in the consensus play an essential role in dimerization. Phylogenetic analysis permitted classification of the 638 bHLH genes identified into 32 subfamilies. Evolutionary and functional relationships within subfamilies are supported by intron patterns, predicted DNA-binding motifs, and the architecture of conserved protein motifs. Our analyses reveal the origin and evolutionary diversification of plant bHLHs through differential expansions, domain shuffling, and extensive sequence divergence. At the functional level, this would translate into different subfamilies evolving specific DNA-binding and protein interaction activities as well as differential transcriptional regulatory roles. Our results suggest a role for bHLH proteins in generating plant phenotypic diversity and provide a solid framework for further investigations into the role carried out in the transcriptional regulation of key growth and developmental processes.
西瓜bHLH转录因子家族基因的鉴定及其在非生物胁迫下的表达分析
DOI:10.16420/j.issn.0513-353x.2015-0886
[本文引用: 1]

利用生物信息学方法,在西瓜测序基因组97103 中共鉴定出96 个bHLH 家族成员,其中有94 个可以被定位到西瓜的11 条染色体上。通过基因结构和结构域序列保守性的预测,发现这些基因的序列长度和内含子数量变化很大,但bHLH 结构域序列比较保守。用拟南芥中39 条已知的bHLH 蛋白序列和西瓜的96 条bHLH 蛋白序列构建系统发育树,结果显示西瓜的bHLH 家族可以进一步被分为11 个亚族。运用荧光定量实时PCR 技术,分析了该家族中21 个基因在西瓜响应非生物胁迫时的表达水平,结果表明,8 个基因受低温胁迫诱导表达,13 个基因受ABA胁迫诱导表达,14 个基因受盐胁迫诱导表达。ClabHLH41在3 种胁迫下表达量均显著增加,说明其在低温、ABA 和盐胁迫应答中可能发挥着重要作用。