咪唑啉酮类除草剂是一种广谱除草剂,其特点是活性高、用量少,喷施剂量只有传统除草剂的1/10,常用于防治一年生禾本科杂草、阔叶杂草和多年生杂草。其机理是通过抑制乙酰羟酸合酶(ALS)活性,影响支链氨基酸的合成,引起杂草代谢紊乱,光合能力下降,植株变黄和生长迟缓,达到抑制杂草生长的目的[5-6]。然而,杂草以外的敏感作物也会受到除草剂的影响,喷施后同样发生光合能力下降[7]。因此,在育种过程中培育抗咪唑啉酮类除草剂品种降低除草剂对作物的影响成为行之有效的途径[8⇓-10]。抗咪唑啉酮类除草剂的农作物品种多通过靶位点的突变来实现,单核苷酸突变引起ALS基因编码的氨基酸发生变化,提高对咪唑啉酮类除草剂抗性;此外,也可以通过减少作物对除草剂的吸收或运输、促进除草剂的解毒以及降低除草剂活性等方式提高作物对咪唑啉酮类除草剂的抗性[11⇓⇓⇓-15]。现有抗咪唑啉酮类除草剂谷子品种主要是通过常规杂交育种方法选育,有关谷子对咪唑啉酮类除草剂的抗性数据仅限于生理指标和田间表型分析[16],关于其抗性的分子机制研究较少。因此,本研究通过高通量RNA测序(RNA-Seq)来挖掘和鉴定咪唑啉酮类除草剂在抗性和敏感谷子品种中差异表达的基因和代谢途径,解析咪唑啉酮类除草剂处理后抗性和敏感性谷子品种表型差异的原因,为后续使用分子技术提高谷子对咪唑啉酮类除草剂的抗性提供基因资源。
1 材料与方法
1.1 试验材料和试验处理
咪唑啉酮类除草剂抗性品种(R)5058和敏感品种(S)豫谷18来源于安阳市农业科学院谷子研究所资源库。咪唑啉酮类除草剂选用德国巴斯夫(中国)有限公司生产的甲咪唑烟酸水剂。在谷子出苗后15 d向谷子幼苗均匀地喷洒甲咪唑烟酸(1125 mL/hm2)进行处理,以清水喷施作对照,设置3个重复,采样时间为处理后0和48 h,取样部位为第3片幼叶,田间取样后放入液氮中带回,样本在-80 ℃冰箱中保存。
1.2 RNA提取、cDNA文库构建和测序
按照说明书的描述,使用RNA提取试剂盒RNAeasyminikit从抗性和敏感品种的幼叶组织中提取总RNA。使用不含RNA酶的DNase去除样品中的基因组DNA。使用NanoPhotometer®分光光度计评估RNA纯度。分别使用RNA分析试剂盒Qubit® RNAAssayKit和AgilentBioanalyzer 2100系统评估RNA浓度和完整性。使用poly-T寡核苷酸附着的磁珠从总RNA中纯化mRNA。按照说明书,使用NEBNext®UltraTMRNALibraryPrepKitfor Illumina®构建测序文库,在IlluminaHiseq 4000平台上对制备的文库进行测序,以生成150 bp的配对末端读数。测序数据保存在美国国家生物技术信息中心(NCBI)(BioProject登录号:PRJNA590609;BioSample登录号:SAMN13335985、SAMN13335986、SAMN13335987、SAMN133359858、SAMN13335989和SAMN13335990)。
1.3 转录组数据的预处理
1.4 差异表达分析
使用R语言的edgeR包识别差异表达基因。2组之间的倍数变化计算为logFC=log2(处理组/对照组)。logFC>1和q<0.05的基因被认为是差异表达基因(DEG)。
1.5 DEG的功能富集与分析
使用TermFinder软件对所有DEG进行GeneOntology(GO,
1.6 实时定量PCR分析
表1 qRT-PCR所用引物
Table 1
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| SiACTIN7-F | GGCAAACAGGGAGAAGATGA |
| SiACTIN7-R | GAGGTTGTCGGTAAGGTCACG |
| LOC101765996-F | CATTCACAGCCTGAGGTGTTTCC |
| LOC101765996-R | CCATCTCCGACATCTCGCATT |
| LOC101784847-F | GACATCCCGGAGGTGCTCAA |
| LOC101784847-R | CGTCAGGCTCGGCATTCAA |
| LOC101759205-F | AGACATCACCGACCTGTTCCAA |
| LOC101759205-R | GCCCAGCACTTGTTCTCACG |
| LOC101765796-F | ACGCCATCAACTTCCCCATC |
| LOC101765796-R | GCCTTGTAGACGACGACCCA |
| LOC101782898-F | AGACAACCGAAAATCAGCAGACAG |
| LOC101782898-R | TGCCCTCAGGTATGCCCAGT |
2 结果与分析
2.1 不同抗性谷子品种的转录组测序概况
本研究共构建了18个cDNA文库,过滤原始读数后有27 620 386~45 776 540个有效数据(表2)。大约88.13%~93.53%的数据映射到谷子参考基因组,82.49%~90.81%的数据可以唯一匹配到谷子参考基因组。18个文库的Q20和Q30分别在96.40%和91.63%以上。
表2 谷子转录组数据
Table 2
| 样本 Sample | 有效数据 Clean reads | 映射数据 Mapped reads | 映射率 Mapped rate (%) | 唯一映射率 Unique mapped rate (%) | GC (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|---|---|
| R0_1 | 35 066 474 | 31 574 750 | 90.04 | 87.36 | 56.00 | 96.67 | 91.78 |
| R0_2 | 27 620 386 | 25 642 394 | 92.84 | 89.43 | 54.00 | 97.23 | 92.65 |
| R0_3 | 30 454 968 | 28 403 582 | 93.26 | 89.87 | 55.50 | 97.36 | 92.86 |
| S0_1 | 32 637 848 | 29 758 054 | 91.18 | 87.40 | 55.00 | 96.75 | 92.06 |
| S0_2 | 32 213 222 | 29 246 420 | 90.79 | 86.69 | 55.00 | 96.76 | 92.11 |
| S0_3 | 30 212 562 | 27 707 582 | 91.71 | 86.30 | 54.50 | 96.91 | 92.26 |
| RT_1 | 41 040 330 | 38 384 804 | 93.53 | 90.81 | 53.50 | 97.48 | 93.26 |
| RT_2 | 32 272 636 | 29 688 540 | 91.99 | 88.97 | 54.00 | 97.19 | 92.74 |
| RT_3 | 31 474 482 | 28 009 464 | 88.99 | 86.24 | 53.50 | 96.55 | 91.90 |
| ST_1 | 31 395 192 | 28 837 638 | 91.85 | 89.42 | 53.00 | 96.82 | 92.31 |
| ST_2 | 32 216 306 | 29 179 166 | 90.57 | 87.82 | 53.50 | 96.92 | 92.42 |
| ST_3 | 38 151 776 | 35 326 060 | 92.59 | 89.61 | 53.00 | 97.26 | 92.91 |
| WRT_1 | 30 168 990 | 26 748 998 | 88.66 | 82.49 | 54.00 | 96.40 | 91.63 |
| WRT_2 | 41 280 980 | 38 641 026 | 93.60 | 89.58 | 55.00 | 97.75 | 93.49 |
| WRT_3 | 33 018 182 | 30 868 352 | 93.49 | 89.25 | 55.00 | 97.66 | 93.26 |
| WST_1 | 42 602 542 | 38 785 788 | 91.04 | 87.82 | 55.50 | 97.80 | 93.64 |
| WST_2 | 38 116 254 | 33 591 880 | 88.13 | 84.78 | 55.50 | 97.78 | 93.60 |
| WST_3 | 45 776 540 | 41 381 022 | 90.40 | 87.53 | 56.50 | 97.85 | 93.66 |
R0表示未处理的抗性品种,S0表示未处理的敏感品种,RT表示除草剂处理的抗性品种,ST表示除草剂处理的抗性品种,WRT表示水处理的抗性品种,WST表示水处理的敏感品种,GC表示鸟嘌呤和胞嘧啶含量,Q20表示质量值≥20的核苷酸的百分比,Q30表示质量值≥30的核苷酸的百分比。
R0 indicates untreated resistant varieties, S0 indicates untreated sensitive varieties, RT indicates herbicide treated resistant varieties, ST indicates herbicide treated resistant varieties, WRT indicates water treated resistant varieties, WST indicates water treated sensitive varieties, GC indicates guanine and cytosine content, Q20 indicates percentage of nucleotides with mass value ≥20. Q30 indicates the percentage of nucleotides with a mass value ≥30.
2.2 不同比较组之间DEG的鉴定
为了鉴定不同基因谷子抗除草剂的关系,比较了不同分组下的基因情况,分组包括除草剂处理48 h的抗性品种与处理0 h的抗性品种比较(RT vs R0)、除草剂处理48 h的抗性品种与水处理48 h的抗性品种比较(RT vs WRT)、水处理48 h的抗性品种与水处理48 h的抗性品种比较(WRT vs R0)、除草剂处理48 h的敏感品种与水处理48 h的敏感品种比较(ST vs WST)、除草剂处理48 h的敏感品种与处理0 h的敏感品种比较(ST vsS0)、水处理48 h的敏感品种与水处理48 h的敏感品种比较(WST vs S0)。与R0相比,RT鉴定出1413个DEG(表3)。其中上调983个,下调430个。此外,与S0相比,ST鉴定出7453个DEG。其中上调3566个,下调3887个。与WRT相比,RT鉴定出743个DEG,其中上调615个,下调128个。与WST相比,ST鉴定出5298个DEG,其中上调2396个,下调2902个。与R0相比,WRT鉴定出186个DEG。其中上调71个,下调115个。与S0相比,WST鉴定出90个DEG。其中上调453个,下调467个。
表3 差异表达基因的数量
Table 3
| 比较组 Comparison group | DEG数 DEG number | 上调 Up-regulated | 下调 Down-regulated |
|---|---|---|---|
| RT vs R0 | 1413 | 983 | 430 |
| ST vs S0 | 7453 | 3566 | 3887 |
| RT vs WRT | 743 | 615 | 128 |
| ST vs WST | 5298 | 2396 | 2902 |
| WRT vs R0 | 186 | 71 | 115 |
| WST vs S0 | 920 | 453 | 467 |
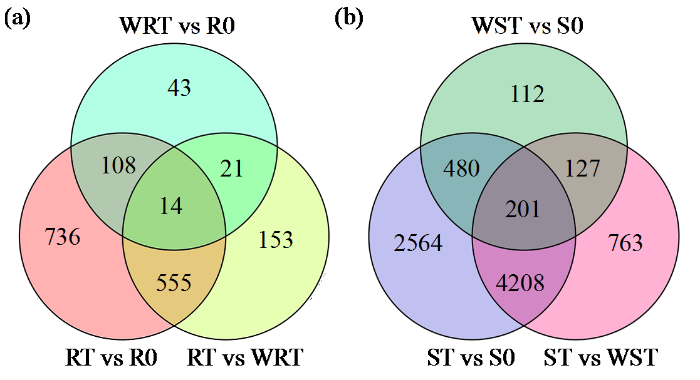
综合比较发现,用甲咪唑烟酸除草剂的处理分别有153个(RT vs WRT)和763个(ST vs WST)DEG在抗性和敏感品种中发生了变化(图1)。
图1
图1
除草剂和水处理0 h和48 h后抗性(a)和敏感(b)品种差异表达基因的维恩图
Fig.1
Venn diagram of differentially expressed genes in resistant (a) and sensitive (b) cultivars under 0 h and 48 h of herbicide or water treatments
2.3 DEG的GO注释分析
图2
图2
处理后谷子差异表达基因的GO分类
Fig.2
GO classifications of foxtail millet differentially expressed genes after treatment
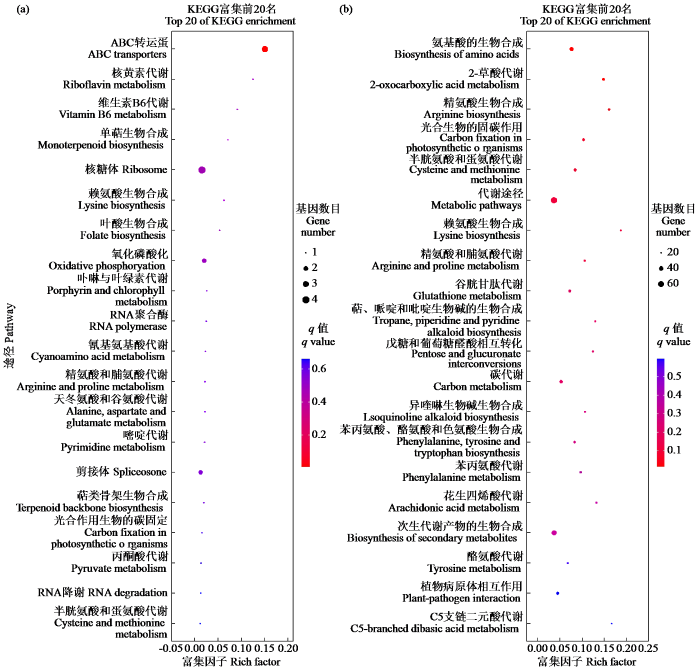
图3
图3
20条KEGG通路显著富集的散点图
(a) RT vs WRT;(b) ST vs WST。富集因子是该通路注释的DEG数与该通路注释的所有基因数的比值。更大的富集因子意味着更大的密集度。q值是修正后的p值,取值范围在0到1之间。q值越低,富集越显著。
Fig.3
Scatter plot of 20 significantly enriched KEGG pathways
(a) RT vs WRT; (b) ST vs WST. Rich factor is the DEG numbers ratio annotated in this pathway term to all gene numbers annotated in this pathway term. Greater rich factor means greater intensiveness. q-value is corrected p-value ranging from 0 to 1. The lower the q value, the more significant the enrichment.
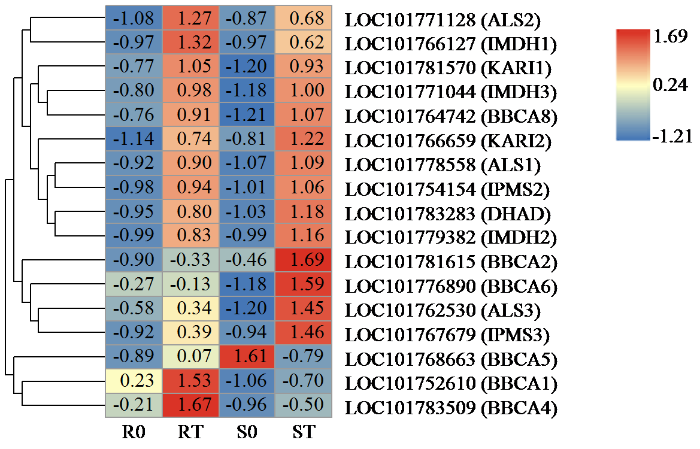
2.4 与除草剂反应差异相关的支链氨基酸合成相关基因
在处理后的2个品种中鉴定出17个与支链氨基酸合成相关的DEG(图4),包括3个乙酰羟酸合酶基因(ALS)、2个酮醇酸还原异构酶(KARI)、1个二羟酸脱水酶(DHAD)、2个2-异丙基苹果酸合酶(IPMS)、3个3-异丙基苹果酸脱氢酶(IMDH)。6个分支氨基酸转氨酶(BBCA)。除草剂处理后,几乎所有的基因在2个品种中都有所提高,其中ALS2、IMDH1、BBCA1和BBCA4在抗性品种中上调更明显。ALS3、IMPS3、BBCA2和BBCA6的表达在敏感品种中上调更明显。总体来说,支链氨基酸合成相关的DEG在2个品种中的表达趋势差异不大。
图4
图4
支链氨基酸合成相关差异表达基因热图分析
条形图表示热图中每个基因表达水平(log2 TPM)。基因的表达水平用不同的颜色表示。红色表示高表达,蓝色表示低表达。
Fig.4
Heatmap analysis of DEGs related to branched chain amino acid synthesis
The bar represents the scale of each gene's expression levels (log2 TPM) in the heat map. The expression level of the genes is denoted using different colours. Red denotes high expression while blue denotes low expression.
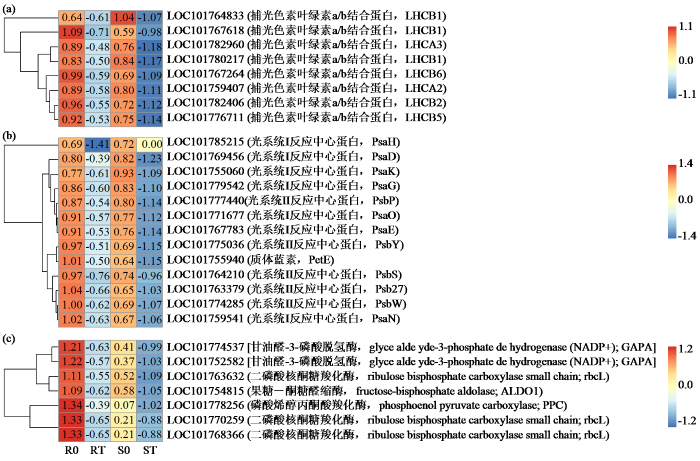
2.5 与除草剂反应差异相关的光合作用相关基因
处理后2个品种均有许多与天线蛋白、光系统和固碳相关的基因(图5)。在处理后的2个品种中鉴定出8个与天线蛋白相关的DEG(图5a),包括2个LHCI天线蛋白和6个LHCII天线蛋白。在2个品种中除草剂处理后这些基因的表达有所下降。然而,它们的表达在敏感品种中比在抗性品种中受到更多抑制。此外,与光系统相关的13个DEG的表达量在敏感品种中也更明显的下调(图5b)。同时,光合作用的暗反应也受到了除草剂的影响。在2个品种除草剂处理后,与碳固定相关的7个DEG的表达均下调。和上述结果一致,这些基因在敏感品种中受到更显著的抑制(图5c)。结果表明,相比于敏感品种,抗除草剂品种在除草剂胁迫下可以保持光合基因相对正常的表达。
图5
图5
光合相关DEG热图分析
(a) 与天线蛋白相关的DEG;(b) 与光系统相关的DEG;(c) 与碳固定相关的DEG。条形图表示热图中每个基因表达水平(log2 TPM)。基因的表达水平用不同的颜色表示。红色表示高表达,蓝色表示低表达。下同。
Fig.5
Heatmap analysis of DEGs related to photosynthesis
(a) DEGs associated with antenna protein; (b) DEGs associated with photosystem; (c) DEGs related to carbon fixation. The bar represents the scale of each gene's expression levels (log2 TPM) in the heat map. The expression level of the genes is denoted using different colours. Red denotes high expression while blue denotes low expression.The same below.
2.6 分析可能与除草剂反应相关的代谢途径基因
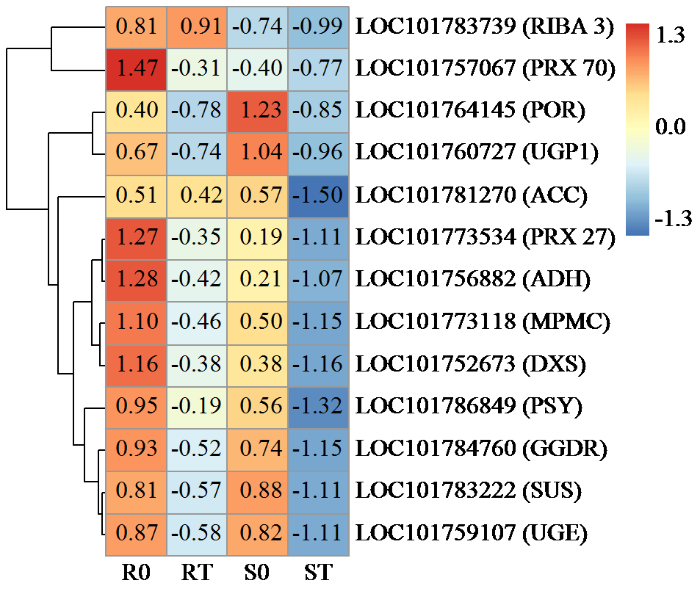
鉴定了与代谢途径相关的13个DEG。它们包括八氢番茄红素合酶(PSY)、香叶基香叶基二磷酸还原酶(GGDR)、单功能核黄素生物合成蛋白(RIBA3)、蔗糖合酶(SUS)、乙酰辅酶A羧化酶(ACC)、过氧化物酶27(PRX27)、过氧化物酶70(PRX70)、镁-原卟啉IX单甲酯[氧化]环化酶(MPMC)、原叶绿素还原酶(POR)、UTP-葡萄糖-1-磷酸尿苷酰转移酶(UGP1)、UDP-葡萄糖4-差向异构酶(UGE)、乙醇脱氢酶(ADH)和1-脱氧-D-木酮糖-5-磷酸合酶(DXS)。除草剂处理后,RIBA3在抗性品种中上调,但在敏感品种中下调(图6)。ACC的表达水平在抗除草剂品种中无明显变化,在敏感品种中显著下调。除草剂处理后,2个品种的其余11个基因均被下调。尽管如此,它们在敏感品种中的表达比在抗性品种中受到更多的抑制。
图6
图6
涉及代谢途径的DEG的热图分析
条形图表示热图中每个基因表达水平(log2 TPM)。
Fig.6
Heatmap analysis of DEGs involved in the metabolic pathway
The bar represents the expression level of each gene (log2 TPM) in the heat map.
2.7 可能与除草剂反应相关的脂肪酸延伸基因分析
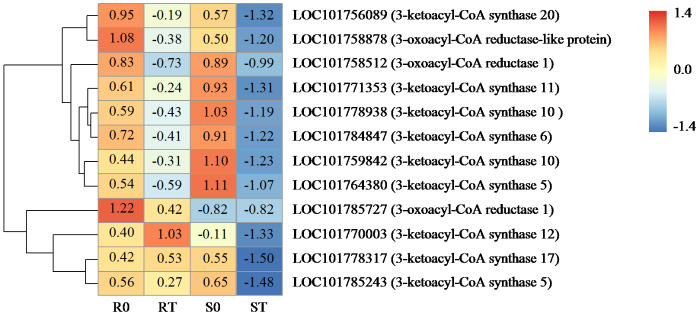
在植物中,3-酮酰基-CoA合酶和3-氧代酰基-CoA还原酶是与超长链脂肪酸(VLCFA)生物合成相关的重要酶。抗除草剂品种的KCS5(LOC101785243)和KCS17(LOC101778317)在处理后的表达水平没有显著变化。然而,这2个基因在敏感品种中表达均显著下调(图7)。此外,KCS5(LOC101764380)、KCS6(LOC101784847)、2个KCS10(LOC101778938和LOC101759842)、KCS11(LOC101771353)、KCS12(LOM101770003)、KCS20(LOC101756089)和3个3-氧代酰基-CoA还原酶基因(LOC101758878,LOC101758878,LOC101785727)在除草剂处理后在2个品种中均下调。然而,这些基因在敏感品种中比在抗性品种中受到更多抑制。结果表明,一些与长链脂肪酸生物合成途径相关基因的表达在2个品种中存在显著差异。
图7
图7
超长链脂肪酸延长酶相关DEG的热图分析
Fig.7
Heatmap analysis of DEGs related to very-long-chain fatty acid elongases
2.8 RNA-Seq测序数据的qRT-PCR验证
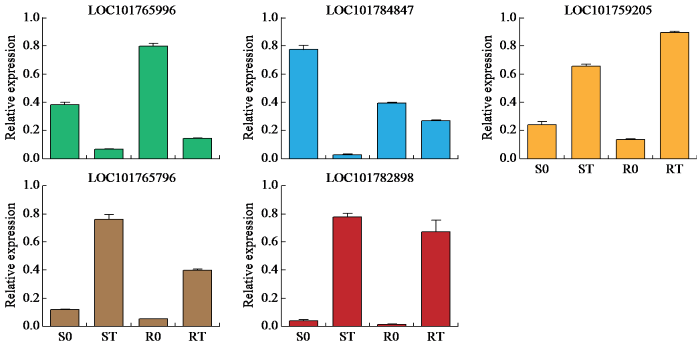
使用qRT-PCR以验证RNA-Seq数据的可靠性。随机选择了5个DEG进行验证,包括LOC101765996(无机磷酸盐转运蛋白)、LOC101784847(3-酮脂酰辅酶A合酶6)、LOC101759205(乙烯响应转录因子)、LOC101765796(蛋白质解毒40)和LOC101782898(ATP结合盒式转运蛋白家族成员15)。它们的表达在除草剂处理后受到影响,与RNA-Seq结果一致,从而证实了RNA-Seq数据的可重复性(图8)。
图8
3 讨论
3.1 除草剂处理后支链氨基酸合成相关基因的表达情况
3.2 与光合作用相关的DEG在敏感品种中显著下调
植物的光合性能对除草剂等胁迫因素很敏感,光合作用及其相关参数是除草剂引起的参考指标之一[5]。前人研究认为,咪唑啉酮类除草剂会引起向日葵叶片萎黄[23],还对向日葵的光合氧化还原反应和叶片气体交换过程产生不利影响[22];甲氧咪草烟处理能降低小麦植株的CO2固定率[6],谷子敏感材料喷施甲咪唑乙烟酸后会显著谷子发芽率和存活率,甚至致死,谷子幼苗在喷药第7天,敏感材料的抗逆生理指标超氧化物歧化酶(SOD)、过氧化物酶(POD)和过氧化氢酶(CAT)均表现显著下降[16]。本研究发现,经咪唑啉酮除草剂处理后在敏感品种和抗性品种中均下调了天线蛋白相关的DEG,包括2个LHCⅠ天线蛋白和6个LHCⅡ天线蛋白,然而这些基因的表达在敏感品种中比在抗性品种中受到更多的抑制;与光合作用相关的13个DEG在敏感品种中也显著下调;在CO2固定阶段,除草剂处理后与碳固定相关的7个DEG在2个品种中均有所下调,敏感品种中比在抗性品种中下调的幅度更明显,这一发现与前人[16]研究结果一致。
3.3 与代谢过程相关的DEG的表达在敏感品种中受到更多抑制
除了破坏植物的光合作用和能量代谢过程外,咪唑啉酮除草剂还会影响作物的一系列代谢过程。许多与代谢过程相关的酶在植物对非生物胁迫的反应中发挥着关键作用。类胡萝卜素是植物抗氧化系统的成员,在清除植物中的活性氧物质、帮助植物抵抗与应激相关的损害方面发挥着至关重要的作用[25],PSY是类胡萝卜素生物合成途径中的一种必需酶,POD负责提高植物的非生物胁迫耐受性[26],ADH赋予植物非生物和生物胁迫抗性。此外,一些酶参与植物的叶绿素合成。单功能核黄素生物合成蛋白、镁―原卟啉IX单甲基酯氧化环化酶和原叶绿素还原酶的缺失会导致黄化幼苗的产生[27]。本研究认为,RIBA3的表达在抗性谷子品种中上调,但在除草剂处理后的敏感品种中下调,抗除草剂品种处理后ACC的表达水平无明显变化,但敏感品种中显著下调。此外,与植物代谢过程相关的其他基因同样在敏感品种中比在抗性品种中受到更多抑制。
3.4 与超长链脂肪酸合成相关的DEG的表达在敏感品种中受到更多抑制
4 结论
谷子抗咪唑啉酮除草剂转录组分析表明,谷子敏感品种和抗性品的差异表达基因影响了谷子对除草剂的抗性,包括与光合作用、代谢途径和脂肪酸延伸有关的基因,表明抗性和敏感的谷子品种之间存在着复杂的抗性机制。
参考文献
Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses
DOI:10.1007/s00122-014-2399-3
PMID:25239219
[本文引用: 1]

Recent advances in Setaria genomics appear promising for genetic improvement of cereals and biofuel crops towards providing multiple securities to the steadily increasing global population. The prominent attributes of foxtail millet (Setaria italica, cultivated) and green foxtail (S. viridis, wild) including small genome size, short life-cycle, in-breeding nature, genetic close-relatedness to several cereals, millets and bioenergy grasses, and potential abiotic stress tolerance have accentuated these two Setaria species as novel model system for studying C4 photosynthesis, stress biology and biofuel traits. Considering this, studies have been performed on structural and functional genomics of these plants to develop genetic and genomic resources, and to delineate the physiology and molecular biology of stress tolerance, for the improvement of millets, cereals and bioenergy grasses. The release of foxtail millet genome sequence has provided a new dimension to Setaria genomics, resulting in large-scale development of genetic and genomic tools, construction of informative databases, and genome-wide association and functional genomic studies. In this context, this review discusses the advancements made in Setaria genomics, which have generated a considerable knowledge that could be used for the improvement of millets, cereals and biofuel crops. Further, this review also shows the nutritional potential of foxtail millet in providing health benefits to global population and provides a preliminary information on introgressing the nutritional properties in graminaceous species through molecular breeding and transgene-based approaches.
谷子bZIP转录因子的全基因组鉴定及其在干旱和盐胁迫下的表达分析
DOI:10.11983/CBB15148
[本文引用: 1]

bZIP蛋白是植物转录因子中最大和最保守的一类转录因子, 参与调控植物生长发育等多种生命活动。谷子(Setaria italica)是一种重要的C4杂粮作物, 其bZIP基因家族与功能报道较少。利用生物信息学工具, 从谷子全基因组中鉴定出73个SibZIP转录因子, 划分为A、B、C、D、E、G、H、I和X等亚家族。与已测序的禾谷类作物相比, 谷子SibZIP基因家族在进化中发生缩减。在谷子SibZIP蛋白中检测到25种不同的保守氨基酸基序。RNA-seq和定量PCR检测结果表明, 在干旱和盐胁迫条件下, 多数SibZIPs基因不同程度地被诱导表达, 预示着部分SibZIP成员在谷子干旱和盐胁迫响应中起重要作用。共表达关联性分析进一步揭示19个谷子SibZIP转录因子可通过与蛋白激酶或NPR1相关调节蛋白等互作介导谷子胁迫响应。研究结果为全面解析谷子SibZIPs基因结构与生物学功能、抗旱分子机制以及分子育种提供了新信息。
Chlorophyll fluorescence as a marker for herbicide mechanisms of action
Resistance to imazamox in Clearfield soft wheat (Triticum aestivum L.)
Glyphosate effect on shikimate, nitrate reductase activity, yield, and seed composition in corn
DOI:10.1021/jf904121y
PMID:20180575
[本文引用: 1]

When glyphosate is applied to glyphosate-resistant (GR) crops, drift to nonglyphosate-resistant (non-GR) crops may cause significant injury and reduce yields. Tools are needed to quantify injury and predict crop losses. In this study, glyphosate drift was simulated by direct application at 12.5% of the recommended label rate to non-GR corn (Zea mays L.) at 3 or 6 weeks after planting (WAP) during two field seasons in the Mississippi delta region of the southeastern USA. Visual plant injury, shikimate accumulation, nitrate reductase activity, leaf nitrogen, yield, and seed composition were evaluated. Effects were also evaluated in GR corn and GR corn with stacked glufosinate-resistant gene at the recommended label rate at 3 and 6 WAP. Glyphosate at 105 g ae/ha was applied once at 3 or 6 weeks after planting to non-GR corn. Glyphosate at 840 (lower label limit) or 1260 (upper label limit) g ae/ha was applied twice at 3 and 6 WAP to transgenic corn. Glyphosate caused injury (45-55%) and increased shikimate levels (24-86%) in non-GR compared to nontreated corn. In non-GR corn, glyphosate drift did not affect starch content but increased seed protein 8-21% while reducing leaf nitrogen reductase activity 46-64%, leaf nitrogen 7-16%, grain yield 49-54%, and seed oil 18-23%. In GR and GR stacked with glufosinate-resistant corn, glyphosate applied at label rates did not affect corn yield, leaf and seed nitrogen, or seed composition (protein, oil, and starch content). Yet, nitrate reductase activity was reduced 5-19% with glyphosate at 840 + 840 g/ha rate and 8-42% with glyphosate at 1260 + 1260 g/ha rate in both GR and GR stacked corn. These results demonstrate the potential for severe yield loss in non-GR corn exposed to glyphosate drift.
Review of glyphosate and als-inhibiting herbicide crop resistance and resistant weed management
Herbicide-resistant crops: Utilities and limitations for herbicide-resistant weed management
DOI:10.1021/jf101286h
PMID:20586458
[本文引用: 1]

Since 1996, genetically modified herbicide-resistant (HR) crops, particularly glyphosate-resistant (GR) crops, have transformed the tactics that corn, soybean, and cotton growers use to manage weeds. The use of GR crops continues to grow, but weeds are adapting to the common practice of using only glyphosate to control weeds. Growers using only a single mode of action to manage weeds need to change to a more diverse array of herbicidal, mechanical, and cultural practices to maintain the effectiveness of glyphosate. Unfortunately, the introduction of GR crops and the high initial efficacy of glyphosate often lead to a decline in the use of other herbicide options and less investment by industry to discover new herbicide active ingredients. With some exceptions, most growers can still manage their weed problems with currently available selective and HR crop-enabled herbicides. However, current crop management systems are in jeopardy given the pace at which weed populations are evolving glyphosate resistance. New HR crop technologies will expand the utility of currently available herbicides and enable new interim solutions for growers to manage HR weeds, but will not replace the long-term need to diversify weed management tactics and discover herbicides with new modes of action. This paper reviews the strengths and weaknesses of anticipated weed management options and the best management practices that growers need to implement in HR crops to maximize the long-term benefits of current technologies and reduce weed shifts to difficult-to-control and HR weeds.
The benefits of herbicide-resistant crops
DOI:10.1002/ps.3374
PMID:22865693
[本文引用: 1]

Since 1996, genetically modified herbicide-resistant crops, primarily glyphosate-resistant soybean, corn, cotton and canola, have helped to revolutionize weed management and have become an important tool in crop production practices. Glyphosate-resistant crops have enabled the implementation of weed management practices that have improved yield and profitability while better protecting the environment. Growers have recognized their benefits and have made glyphosate-resistant crops the most rapidly adopted technology in the history of agriculture. Weed management systems with glyphosate-resistant crops have often relied on glyphosate alone, have been easy to use and have been effective, economical and more environmentally friendly than the systems they have replaced. Glyphosate has worked extremely well in controlling weeds in glyphosate-resistant crops for more than a decade, but some key weeds have evolved resistance, and using glyphosate alone has proved unsustainable. Now, growers need to renew their weed management practices and use glyphosate with other cultural, mechanical and herbicide options in integrated systems. New multiple-herbicide-resistant crops with resistance to glyphosate and other herbicides will expand the utility of existing herbicide technologies and will be an important component of future weed management systems that help to sustain the current benefits of high-efficiency and high-production agriculture.Copyright © 2012 Society of Chemical Industry.
Contribution of non-target-site resistance in imidazolinones-resistant Imisun sunflower
Glutathione transferases in wheat (Triticum) species with activity toward fenoxaprop-ethyl and other herbicides
Glutathione transferase activity and herbicide selectivity in maize and associated weed species
Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds
Engineering the meta- bolism of the phenylurea herbicide chlortoluron in genetically modified Arabidopsis thaliana plants expressing the mammalian cytochrome P450 enzyme CYP1A2
谷子抗咪唑啉酮的遗传应用和基因初定位
DOI:10.11924/j.issn.1000-6850.casb2020-0749
[本文引用: 3]

探究咪唑啉酮除草剂在谷子应用上的安全剂量和最大剂量,并对抗咪唑啉酮进行基因初定位,以期为抗咪唑啉酮谷子育种的利用提供理论基础。以谷子抗性材料‘安5158’和敏感材料‘豫谷9号’为研究对象,分别对其种子和幼苗喷施不同浓度咪唑啉酮,并选用250 mL/hm<sup>2</sup>浓度对亲本和BC<sub>1</sub>群体喷施,结合转录组分析完成抗咪唑啉酮基因的初定位。研究结果表明,在250 mL/hm<sup>2</sup>浓度下,敏感材料‘豫谷9号’只有14%的种子能正常发芽但无法存活,而抗性材料‘安5158’的种子发芽率和存活率均不受影响;敏感材料‘豫谷9号’幼苗在喷药第7天,250 mL/hm<sup>2</sup>浓度下的抗逆生理指标(SOD、POD、CAT、MDA)出现明显的变化,并且不同喷药天数之间差异显著,而抗性材料‘安5158’在450 mL/hm<sup>2</sup>浓度下才表现相似变化趋势。用250 mL/hm<sup>2</sup>浓度咪唑啉酮对亲本和BC<sub>1</sub>群体喷施,回交后代出现50%左右死亡率,符合完全显性遗传规律。基于转录组测序分析,谷子抗咪唑啉酮除草剂基因定位在第1条染色体短臂6.8~30.8 Mb区间内,共有9个与抗咪唑啉酮相关的候选基因。本试验确定了谷子喷施咪唑啉酮除草剂的安全剂量是250 mL/hm<sup>2</sup>,验证了其完全显性遗传规律,并完成抗咪唑啉酮相关候选基因的初定位,为抗除草剂分子辅助育种提供理论基础。
Cutadapt removes adapter sequences from high- throughput sequencing reads
HISAT: a fast spliced aligner with low memory requirements
DOI:10.1038/nmeth.3317
PMID:25751142
[本文引用: 1]

HISAT (hierarchical indexing for spliced alignment of transcripts) is a highly efficient system for aligning reads from RNA sequencing experiments. HISAT uses an indexing scheme based on the Burrows-Wheeler transform and the Ferragina-Manzini (FM) index, employing two types of indexes for alignment: a whole-genome FM index to anchor each alignment and numerous local FM indexes for very rapid extensions of these alignments. HISAT's hierarchical index for the human genome contains 48,000 local FM indexes, each representing a genomic region of ∼64,000 bp. Tests on real and simulated data sets showed that HISAT is the fastest system currently available, with equal or better accuracy than any other method. Despite its large number of indexes, HISAT requires only 4.3 gigabytes of memory. HISAT supports genomes of any size, including those larger than 4 billion bases.
TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions
DOI:10.1186/gb-2013-14-4-r36
PMID:23618408
[本文引用: 1]

TopHat is a popular spliced aligner for RNA-sequence (RNA-seq) experiments. In this paper, we describe TopHat2, which incorporates many significant enhancements to TopHat. TopHat2 can align reads of various lengths produced by the latest sequencing technologies, while allowing for variable-length indels with respect to the reference genome. In addition to de novo spliced alignment, TopHat2 can align reads across fusion breaks, which can occur after genomic translocations. TopHat2 combines the ability to identify novel splice sites with direct mapping to known transcripts, producing sensitive and accurate alignments, even for highly repetitive genomes or in the presence of pseudogenes. TopHat2 is available at http://ccb.jhu.edu/software/tophat.
Analysis of relative gene expression data using real-time quantitative PCR and the method
Genetic analysis of pathway regulation for enhancing branched-chain amino acid biosynthesis in plants
A regulatory hierarchy of the arabidopsis branched-chain amino acid metabolic network
Precision phenotyping of imidazolinones-induced chlorosis in sunflower
DOI:10.1270/jsbbs.64.416
PMID:25914598
[本文引用: 1]

Chlorosis level is a useful parameter to assess imidazolinone resistance in sunflower (Helianthus annuus L.). The aim of this study was to quantify chlorosis through two different methods in sunflower plantlets treated with imazapyr. The genotypes used in this study were two inbred lines reported to be different in their resistance to imidazolinones. Chlorosis was evaluated by spectrophotometrical quantification of photosynthetic leaf pigments and by a bioinformatics-based color analysis. A protocol for pigment extraction was presented which improved pigment stability. Chlorophyll amount decreased significantly when both genotypes were treated with 10 μM of imazapyr. Leaf color was characterized using Tomato Analyzer(®) color test software. A significant positive correlation between color reduction and chlorophyll concentration was found. It suggests that leaf color measurement could be an accurate method to estimate chlorosis and infer chlorophyll levels in sunflower plants. These results highlight a strong relationship between imidazolinone-induced chlorosis and variations in leaf color and in chlorophyll concentration. Both methods are quantitative, rapid, simple, and reproducible. Thus, they could be useful tools for phenotyping and screening large number of plants when breeding for imidazolinone resistance in this species.
Photosynthetic performance of the imidazolinones resistant sunflower exposed to single and combined treatment by the herbicide imazamox and an amino acid extract
PMID:27826304

The herbicide imazamox may provoke temporary yellowing and growth retardation in IMI-R sunflower hybrids, more often under stressful environmental conditions. Although, photosynthetic processes are not the primary sites of imazamox action, they might be influenced; therefore, more information about the photosynthetic performance of the herbicide-treated plants could be valuable for a further improvement of the Clearfield technology. Plant biostimulants have been shown to ameliorate damages caused by different stress factors on plants, but very limited information exists about their effects on herbicide-stressed plants. In order to characterize photosynthetic performance of imazamox-treated sunflower IMI-R plants, we carried out experiments including both single and combined treatments by imazamox and a plant biostimulants containing amino acid extract. We found that imazamox application in a rate of 132 μg per plant (equivalent of 40 g active ingredient ha) induced negative effects on both light-light dependent photosynthetic redox reactions and leaf gas exchange processes, which was much less pronounced after the combined application of imazamox and amino acid extract.
A kaleidoscope of carotenoids
Genes encoding plant-specific class III peroxidases are responsible for increased cold tolerance of the brassinosteroid-insensitive 1mutant
Deficiency in riboflavin biosynthesis affects tetrapyrrole biosynthesis in etiolated Arabidopsis tissue
Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation
Molecular caliper mechanism for determining very-long chain fatty acid length