作物杂志,2024, 第3期: 13–22 doi: 10.16035/j.issn.1001-7283.2024.03.003
不同谷子品种喷施咪唑啉酮除草剂后的转录组分析
宋慧1( ), 王涛2, 邢璐1, 刘俊芳1, 张扬1, 刘金荣1, 陈红旗1(
), 王涛2, 邢璐1, 刘俊芳1, 张扬1, 刘金荣1, 陈红旗1( ), 冯佰利3
), 冯佰利3
- 1安阳市农业科学院,455000,河南安阳
2安阳工学院生物与食品工程学院,455000,河南安阳
3西北农林科技大学农学院/旱区作物逆境生物学国家重点实验室,712100,陕西杨凌
Transcriptome Analysis of Different Foxtail Millet (Setaria italica L.) Varieties Treated with Imazapic Herbicide
Song Hui1( ), Wang Tao2, Xing Lu1, Liu Junfang1, Zhang Yang1, Liu Jinrong1, Chen Hongqi1(
), Wang Tao2, Xing Lu1, Liu Junfang1, Zhang Yang1, Liu Jinrong1, Chen Hongqi1( ), Feng Baili3
), Feng Baili3
- 1Anyang Academy of Agricultural Sciences, Anyang 455000, Henan, China
2College of Biology and Food Engineering, Anyang Institute of Technology, Anyang 455000, Henan, China
3College of Agriculture, Northwest Agriculture and Forestry University / State Key Laboratory of Crop Stress Biology in Arid Areas, Yangling 712100, Shaanxi, China
摘要:
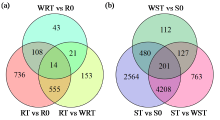
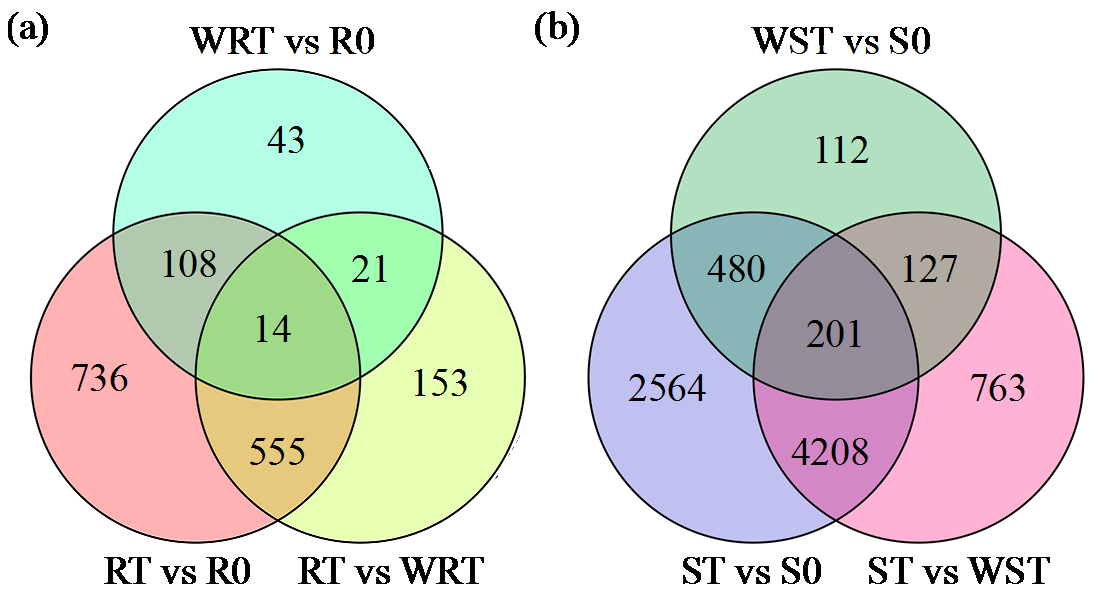
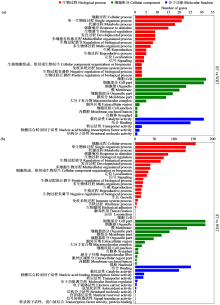
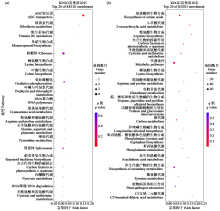
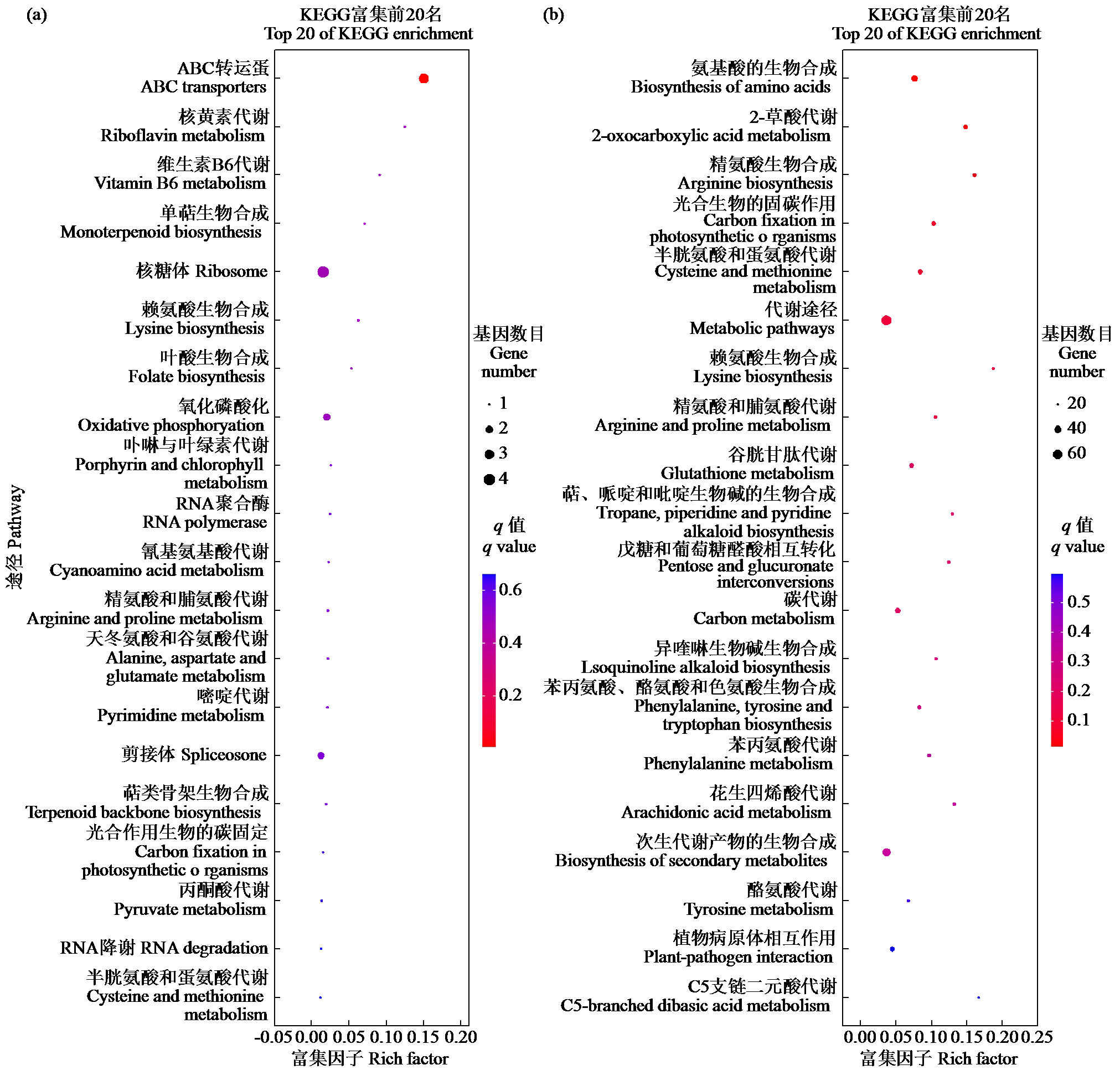
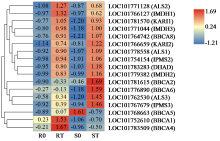
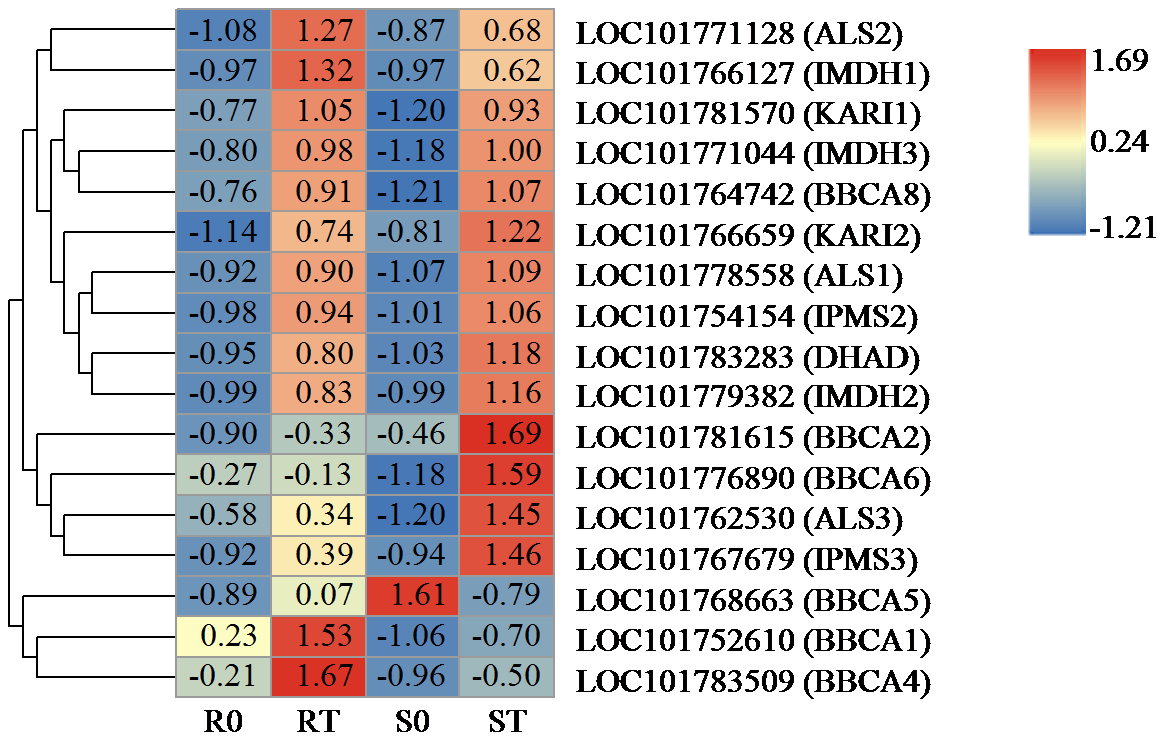
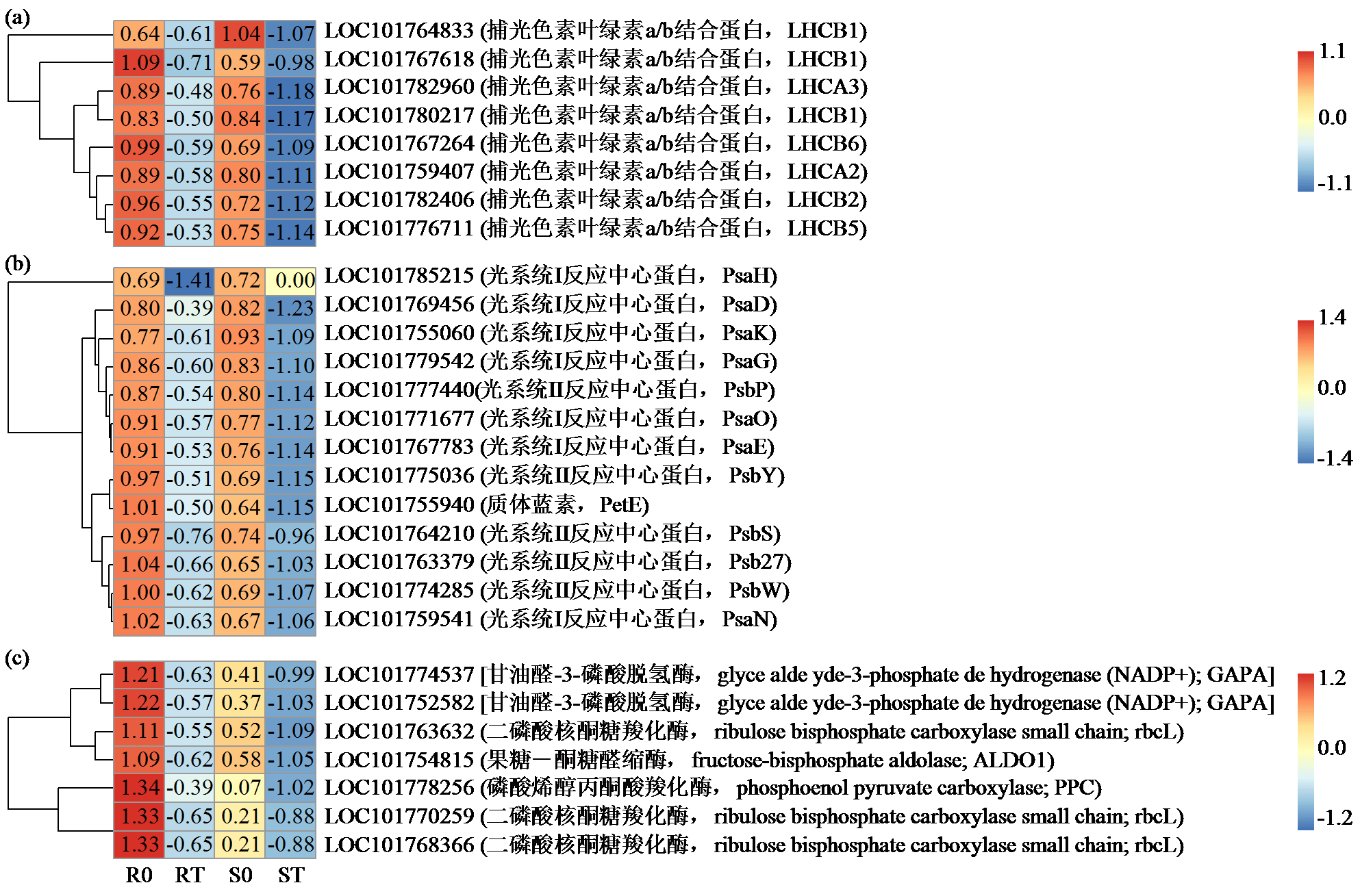
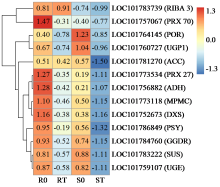
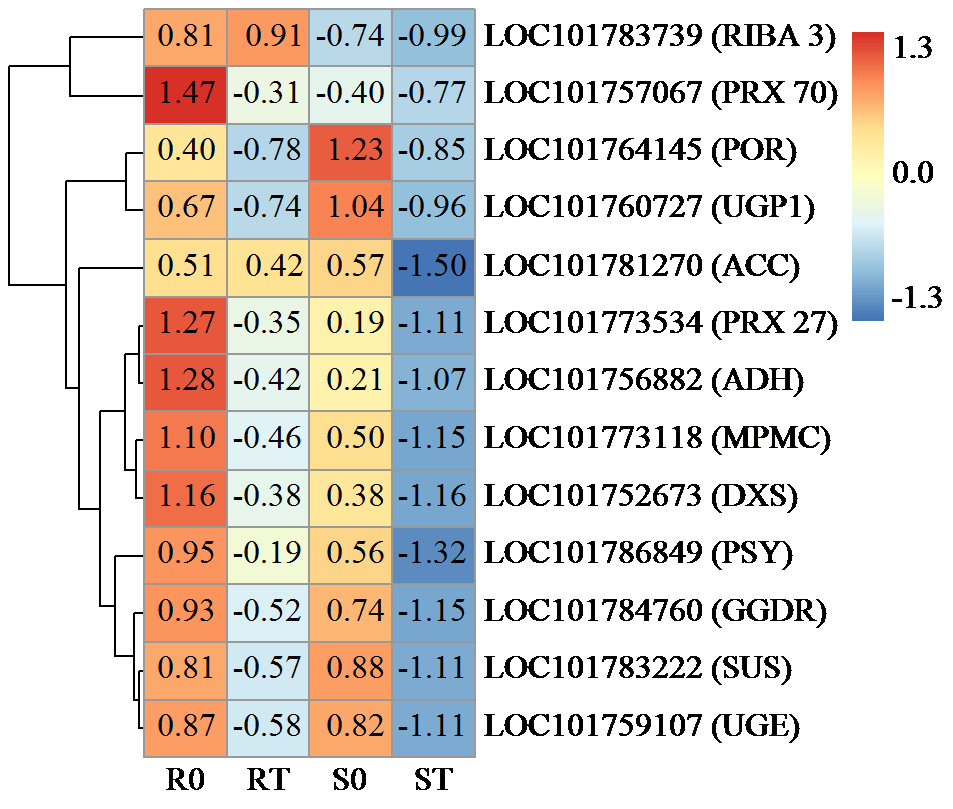
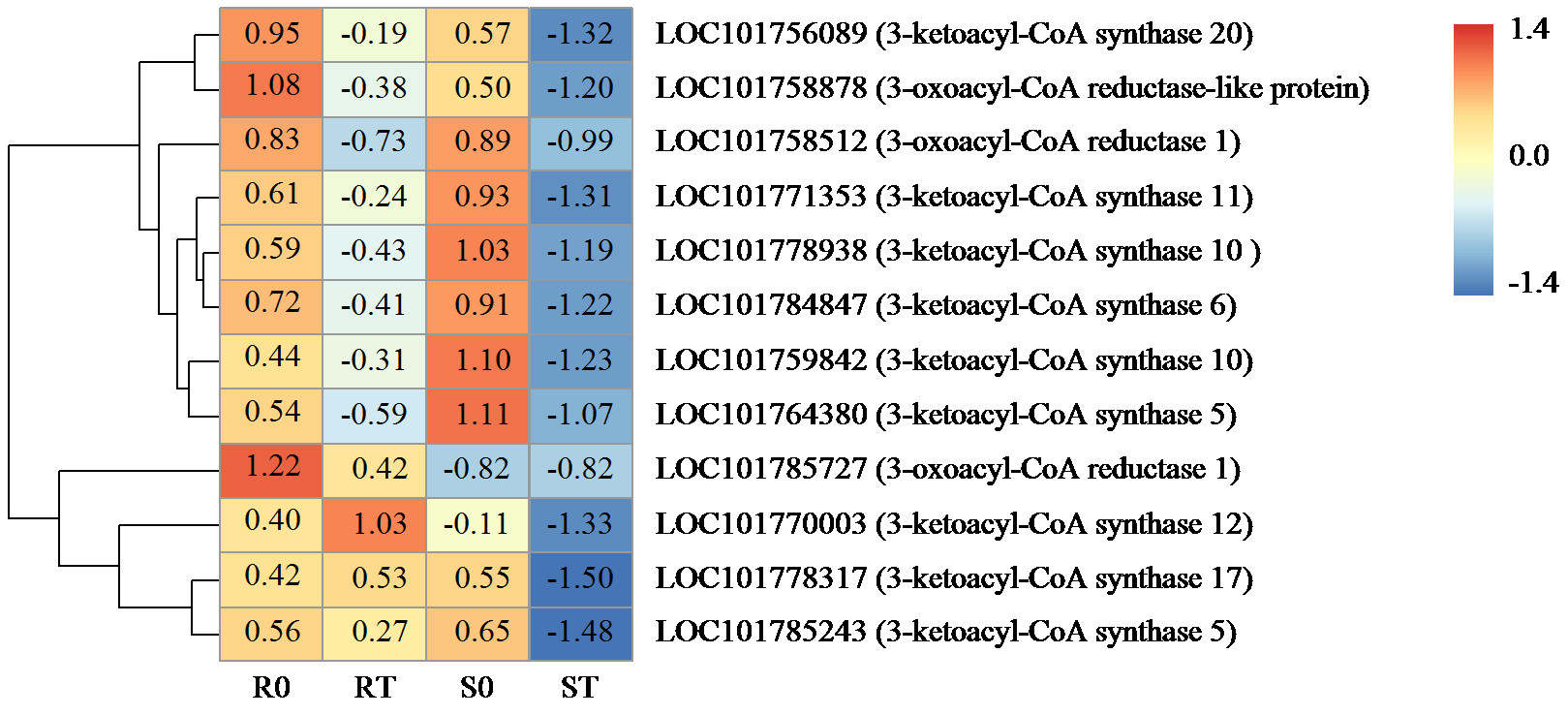
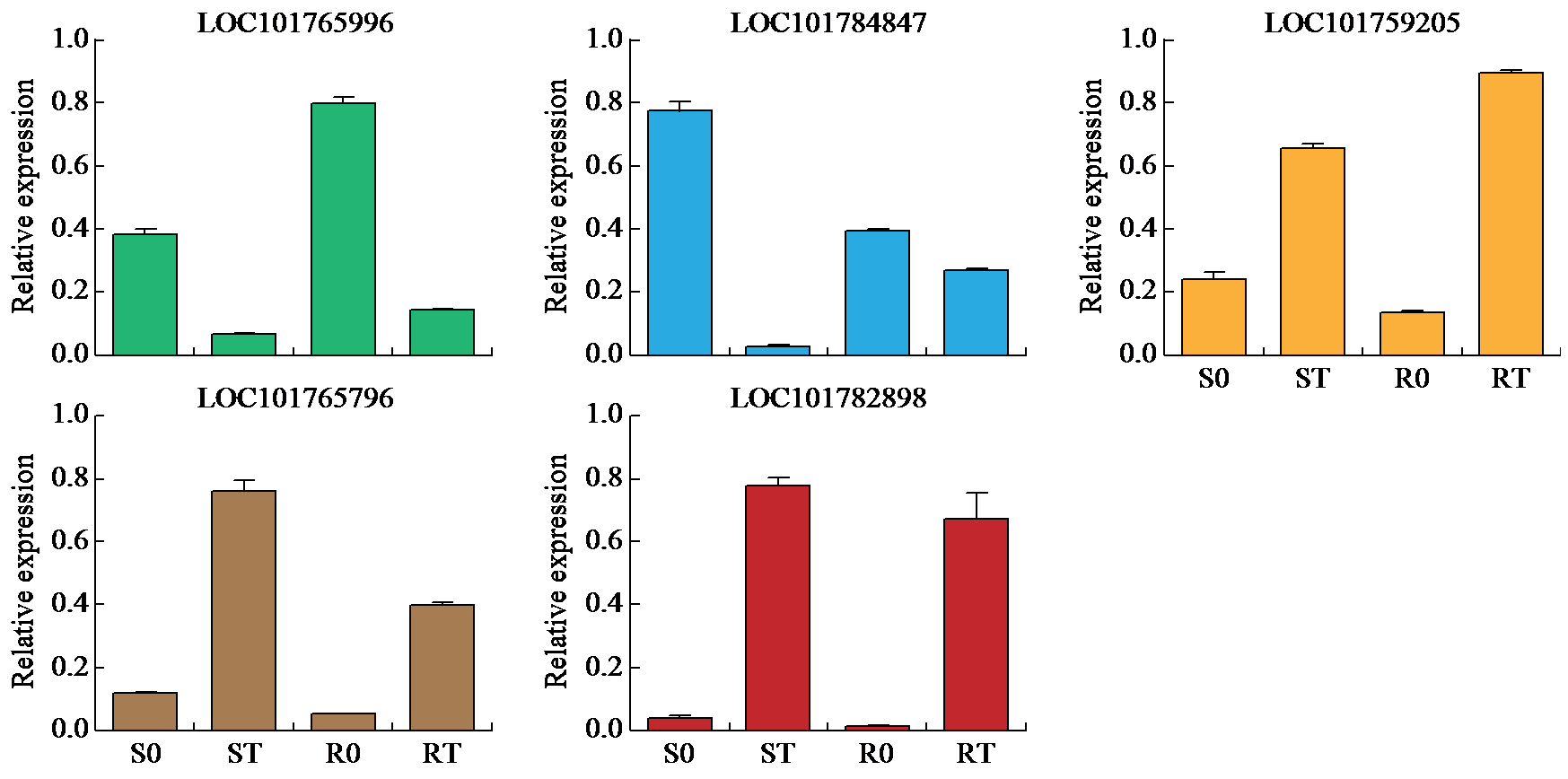
咪唑啉酮类除草剂能有效防治谷子田的单、双子叶杂草,为探究谷子对咪唑啉酮类除草剂抗性的分子机制,于抗性品种(R)和敏感品种(S)出苗后15 d均匀地喷洒甲咪唑烟酸,通过高通量RNA-Seq测序分析抗性和敏感谷子品种中差异表达的基因和代谢途径。结果表明,2个品种在除草剂处理后,与光合作用和代谢途径相关的基因均被下调,特别是在敏感品种中下调更为显著。同样,敏感品种在除草剂处理后,与脂肪酸延伸相关基因表达也显著下调。5个随机选择基因的实时定量PCR(qRT-PCR)的结果与深度测序的结果一致。上述相关基因可能在谷子对咪唑啉酮类除草剂的抗性中起重要作用。
| [1] | 刁现民. 基础研究提升传统作物谷子和黍稷的科研创新水平. 中国农业科学, 2016, 49(17):3260-3262. |
| [2] |
Muthamilarasan M, Prasad M. Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theoretical and Applied Genetics, 2015, 128(1):1-14.
doi: 10.1007/s00122-014-2399-3 pmid: 25239219 |
| [3] |
刘宝玲, 张莉, 孙岩, 等. 谷子bZIP转录因子的全基因组鉴定及其在干旱和盐胁迫下的表达分析. 植物学报, 2016, 51(4):473-487.
doi: 10.11983/CBB15148 |
| [4] | 周汉章, 王新玉, 薄奎勇, 等. 夏谷田阔叶杂草密度与谷子产量损失关系的研究. 作物杂志, 2013(1):108-112. |
| [5] | Dayan F E, Zaccaro M L D M. Chlorophyll fluorescence as a marker for herbicide mechanisms of action. Pesticide Biochemistry and Physiology, 2012, 102:189-197. |
| [6] | Jimenez F, Fernandez P, Rojano-Delgado A M, et al. Resistance to imazamox in Clearfield soft wheat (Triticum aestivum L.). Crop Protection, 2015, 78:15-19. |
| [7] |
Reddy K N, Bellaloui N, Zablotowicz R M. Glyphosate effect on shikimate, nitrate reductase activity, yield, and seed composition in corn. Journal of Agricultural and Food Chemistry, 2010, 58(6):3646-3650.
doi: 10.1021/jf904121y pmid: 20180575 |
| [8] | Green J M. Review of glyphosate and als-inhibiting herbicide crop resistance and resistant weed management. Weed Technology, 2007, 21(2):547-558. |
| [9] |
Green J M, Owen M D K. Herbicide-resistant crops: Utilities and limitations for herbicide-resistant weed management. Journal of Agricultural and Food Chemistry, 2011, 59(11):5819-5829.
doi: 10.1021/jf101286h pmid: 20586458 |
| [10] |
Green J M. The benefits of herbicide-resistant crops. Pest Management Science, 2012, 68(10):1323-1331.
doi: 10.1002/ps.3374 pmid: 22865693 |
| [11] | Breccia G, Gil M, Vega T, et al. Contribution of non-target-site resistance in imidazolinones-resistant Imisun sunflower. Bragantia, 2017, 76(4):536-542. |
| [12] | Edwards R, Cole D J. Glutathione transferases in wheat (Triticum) species with activity toward fenoxaprop-ethyl and other herbicides. Pesticide Biochemistry and Physiology, 1996, 54:96-104. |
| [13] | Hatton P J, Dixon D, Cole D J, et al. Glutathione transferase activity and herbicide selectivity in maize and associated weed species. Pest Management Science, 1996, 46:267-275. |
| [14] | Cummins I, Wortley D J, Sabbadin F, et al. Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(15):5812-5817. |
| [15] | Kebeish R, Azab E, Peterhaensel C, et al. Engineering the meta- bolism of the phenylurea herbicide chlortoluron in genetically modified Arabidopsis thaliana plants expressing the mammalian cytochrome P450 enzyme CYP1A2. Environmental Science and Pollution Research, 2014, 21:8224-8232. |
| [16] |
宋慧, 王涛, 田礼新, 等. 谷子抗咪唑啉酮的遗传应用和基因初定位. 中国农学通报, 2021, 37(28):1-8.
doi: 10.11924/j.issn.1000-6850.casb2020-0749 |
| [17] | Martin M. Cutadapt removes adapter sequences from high- throughput sequencing reads. Embnet Journal, 2011, 17(1):10-12. |
| [18] |
Kim D, Langmead B, Salzberg S L. HISAT: a fast spliced aligner with low memory requirements. Nature Methods, 2015, 12(4):357-360.
doi: 10.1038/nmeth.3317 pmid: 25751142 |
| [19] |
Kim D, Pertea G, Trapnell C, Pimentel H, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 2013, 14(4):36.
doi: 10.1186/gb-2013-14-4-r36 pmid: 23618408 |
| [20] | Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the method. Methods in Molecular Biology, 2011, 25:402-408. |
| [21] | Chen H, Saksa K, Zhao F, et al. Genetic analysis of pathway regulation for enhancing branched-chain amino acid biosynthesis in plants. Plant Journal, 2010, 63(4):573-583. |
| [22] | Xing A, Last R L. A regulatory hierarchy of the arabidopsis branched-chain amino acid metabolic network. Plant Cell, 2017, 29(6):1480-1499. |
| [23] |
Ochogavía A C, Gil M, Picardi L, et al. Precision phenotyping of imidazolinones-induced chlorosis in sunflower. Breeding Science, 2014, 64:416-421.
doi: 10.1270/jsbbs.64.416 pmid: 25914598 |
| [24] |
Balabanova D A, Paunov M, Goltsev V, et al. Photosynthetic performance of the imidazolinones resistant sunflower exposed to single and combined treatment by the herbicide imazamox and an amino acid extract. Frontiers in Plant Science, 2016, 7:1559.
pmid: 27826304 |
| [25] |
Jez J M, Noel J P. A kaleidoscope of carotenoids. Nature Biotechnology, 2000, 18(8):825-826.
pmid: 10932147 |
| [26] | Kim B H, Kim S Y, Nam K H. Genes encoding plant-specific class III peroxidases are responsible for increased cold tolerance of the brassinosteroid-insensitive 1mutant. Molecules & Cells, 2012, 34(6):539-548. |
| [27] | Hedtke B, Alawady A, Albacete A, et al. Deficiency in riboflavin biosynthesis affects tetrapyrrole biosynthesis in etiolated Arabidopsis tissue. Plant Molecular Biology, 2012, 78(1/2):77-93. |
| [28] | Qin Y M, Hu CY, Pang Y, et al. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation. Plant Cell, 2007, 19(11):3692-3704. |
| [29] | Denic V, Weissman J S. Molecular caliper mechanism for determining very-long chain fatty acid length. Cell, 2007, 130:663-677. |
| [1] | 杜杰, 苏福丽, 夏清, 智慧, 王文霞. 谷子苗期对低温胁迫的生理生化响应[J]. 作物杂志, 2024, (3): 180–185 |
| [2] | 包雪莲, 文峰, 金晓光, 呼瑞梅, 黄前晶, 张桂华, 齐金全, 白颖哲, 乌月汗, 白乙拉图. 蒙东粮食主产区不同谷子品种的适应性分析[J]. 作物杂志, 2024, (3): 201–206 |
| [3] | 刘莹, 尹泽群, 吴柏辰, 徐明丽, 刘畅, 石慧姝, 庞博, 苗兴芬. 复合盐碱胁迫对不同谷子品种萌发期的影响及耐盐碱品种筛选[J]. 作物杂志, 2024, (3): 207–215 |
| [4] | 卿晨, 刘正学, 李彦杰. 转录组测序分析干旱胁迫下复合微生物菌肥对玉米幼苗抗旱性的影响[J]. 作物杂志, 2024, (3): 32–39 |
| [5] | 窦维泽, 姜雯, 冯艺川, 金卓, 全雪丽, 吴松权. 过氧化氢诱导膜荚黄芪不定根毛蕊异黄酮葡萄糖苷积累的转录组分析[J]. 作物杂志, 2024, (1): 48–56 |
| [6] | 董好胜, 王琦, 闫鹏, 许艳丽, 张薇, 卢霖, 董志强. 乙矮合剂对谷子茎秆抗倒伏能力及产量的影响[J]. 作物杂志, 2023, (6): 181–189 |
| [7] | 赵丽洁, 赵海燕, 韩根兰, 王江, 聂萌恩, 杜慧玲, 原向阳, 董淑琦. 氮肥配施有机肥对谷子品质的影响[J]. 作物杂志, 2023, (6): 224–232 |
| [8] | 申甜雨, 王媛, 董二伟, 王劲松, 刘秋霞, 焦晓燕. 氮和延迟收获对谷子产量及小米品质的影响[J]. 作物杂志, 2023, (6): 233–242 |
| [9] | 朱文娟, 任月梅, 杨忠, 郭瑞锋, 张绶, 任广兵. 谷子土壤微生物群落结构及功能预测分析[J]. 作物杂志, 2023, (5): 170–178 |
| [10] | 依兵, 刘金刚, 宋殿秀, 王德兴, 赵明珠, 刘晓宏, 孙恩玉, 崔良基. 干旱地区向日葵与谷子间作的土地生产力及种间竞争力的研究[J]. 作物杂志, 2023, (5): 219–223 |
| [11] | 赵海燕, 赵丽洁, 韩根兰, 王江, 王子建, 聂萌恩, 杜慧玲, 原向阳, 董淑琦. 氮锌配施对谷子根系形态及锌含量的影响[J]. 作物杂志, 2023, (4): 152–158 |
| [12] | 刘松涛, 田再民, 刘子刚, 高志佳, 张静, 贺东刚, 黄智鸿, 兰鑫. 基于转录组测序揭示玉米抗倒伏相关基因和代谢通路[J]. 作物杂志, 2023, (4): 31–37 |
| [13] | 赵云, 冯国郡, 胡相伟, 吾买尔江·库尔班, 李鹏兵, 李翠梅, 阿克博塔·木合亚提. 新疆喀什地区适栽抗除草剂复播谷子品种筛选初报[J]. 作物杂志, 2023, (3): 126–133 |
| [14] | 贺水玲, 赵霞, 吴明琦, 王东胜. 外源NO和H2S对谷子种子萌发的影响[J]. 作物杂志, 2023, (2): 138–144 |
| [15] | 马继钰, 王爽, 李云, 郭振清, 王健, 林小虎, 韩玉翠. 种植密度对谷子农艺性状及产量的影响[J]. 作物杂志, 2023, (2): 222–228 |
|
||