植物脂肪酸合成是一个复杂的生物学过程,由溶血磷脂酸(LPA)向磷脂酸(PA)的转化是脂肪酸合成的一个重要的步骤,PA经脱磷酸后生成三酰甘油(TAG),TAG不仅是植物脂质的重要成分,还参与生物膜的形成以及信号转导[4⇓-6]。溶血磷脂酰胆碱酰基转移酶(lysophosphatidylcholine acyltransferase,LPAT)又称溶血磷脂酸酰基转移酶(lysophosphatidic acid acyltransferase,LPAAT)或酰基甘油磷酸酰基转移酶(acylglycerolphosphate acyltransferases,AGPAT),是上述途径的一个关键限速酶,在植物脂肪酸的积累和代谢中起着至关重要的作用[7-8]。异源过表达BnLPAT增加了拟南芥种子含油量[9];在酵母中表达三角褐指藻LPAAT基因明显提高了脂肪酸含量[10]。截至目前,LPAAT基因在许多植物中均被鉴定到,包括拟南芥(Arabidopsis thaliana)[11]、棉花(Gossypium raimondii)[12]、花生(Arachis hypogaea)[13]、紫苏(Perilla frutescens)[14]等。
1 材料与方法
1.1 供试植物材料与处理
甘蓝型油菜中双11(ZS11,高含油量)、绵油16(MY16,低含油量)和绵油32(MY32,中含油量)于2022-2023年种植在四川省绵阳市农业科学研究院内试验基地。分别于花后(days after flowering,DAF)0(花序)、7、14、21、28、35、42 d收集中双11不同发育阶段的种子,绵油16、绵油32的种子于DAF 28和35 d收集,立即放入液氮中速冻,之后转移到-80 ℃超低温冰箱中保存备用。
1.2 LPAT家族成员鉴定与理化性质分析
甘蓝型油菜中双11(Brana_ZS_V2.0/)、白菜(Brapa_genome_v3.0)以及甘蓝(Boleraceacapitata_ 446_v1.0)数据库下载自网站BRAD(
1.3 系统发育与染色体定位
1.4 基因结构、保守基序和多序列比对
将甘蓝型油菜LPAT基因的编码序列(CDS)、gDNA序列提交到GSDS 2.0(
1.5 顺式作用元件分析
将甘蓝型油菜LPAT基因上游2000 bp序列提交到PlantCARE数据库(
1.6 转录组数据获取以及qRT-PCR分析
在BnTIR数据库(
表1 试验所用引物
Table 1
| 基因名称Gene name | 上游引物(5′-3′)Forward primer (5′-3′) | 下游引物(5′-3′)Reverse primer (5′-3′) |
|---|---|---|
| BnLPAT1-A | AGCGTTGGATGCATCTGGTTC | GGTTATTGTCAGTTCCTTGA |
| BnLPAT1-C | GCACAATATTAGAGAGTGTT | GGTTATTGTCAGTTCCTTGA |
| BnLPAT4-A | ATGGCAGCAGCAGTGATTGT | GGTCGAACGAGGACATAGCA |
| BnLPAT4-C | ATGGCAGCAGCAGCAGTGAT | CAGAGGTCGAATGAGGACAT |
| BnLPAT12-A | TGAGGCTGATCTTCCAGCATT | AGCAGAACACTCTGCATCGCT |
| BnLPAT14-A | GTATCATATATCAGTGGGCA | ACTGTGCCAGATACACCCTT |
| BnLPAT14-C | TGTATCATATGTCTGCTTCT | ACTGTGCCAGATACACCCTT |
| BnLPAT19-A | AGTATTTGCGGATAATGAGAC | AACCTGACCTCTGAGCGAGA |
| BnLPAT19-C | AAGTGTTTGCTGATAATGAG | ACCTGATCTCTGAGCCAGAAT |
| BnLPAT20-A | TCACATCCTCCATATTATAG | TCAAGTTCGAAGAACGAGCA |
| BnLPAT20-C | CATCCTCCTCCATATTATAGC | AGAACGACCAAAGCAACATC |
| BnLAPT22-C | TGTATCATATGTCTGCTTCT | CCTCTCGTTGGACATAGACA |
| BnActin7 | TGGGTTTGCTGGTGACGAT | TGCCTAGGACGACCAACAATACT |
2 结果与分析
2.1 BnLPAT家族成员鉴定与理化性质分析
表2 甘蓝型油菜BnLPAT蛋白理化性质和亚细胞定位
Table 2
| 基因编号 Gene ID | 基因名称 Gene name | 氨基酸数目 Number of amino acids | 分子质量 Molecular weight (kDa) | 理论 等电点 pI | 不稳定指数 Instability index | 脂肪系数 Aliphatic coefficient | 水溶性平均值 Average water solubility | 亚细胞定位 Subcellular localization | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BnA01g0003370.1 | BnLPAT1-A | 361 | 41.53 | 8.93 | 46.58 | 87.20 | -0.170 | 叶绿体,内质网 | |||||
| BnA01g0033510.1 | BnLPAT2-A | 355 | 41.02 | 8.77 | 39.55 | 94.96 | -0.019 | 内质网 | |||||
| BnA03g0114510.1 | BnLPAT3-A | 527 | 59.81 | 6.15 | 38.63 | 90.61 | -0.050 | 线粒体 | |||||
| BnA03g0128320.1 | BnLPAT4-A | 390 | 43.77 | 9.24 | 50.09 | 97.26 | 0.067 | 内质网 | |||||
| BnA03g0133720.1 | BnLPAT5-A | 432 | 48.25 | 9.63 | 45.85 | 93.87 | -0.224 | 内质网 | |||||
| BnA03g0144790.1 | BnLPAT6-A | 286 | 32.09 | 9.13 | 52.32 | 96.12 | -0.041 | 叶绿体,内质网 | |||||
| BnA04g0162010.1 | BnLPAT7-A | 384 | 43.19 | 9.18 | 41.28 | 101.59 | 0.136 | 内质网 | |||||
| BnA05g0207140.1 | BnLPAT8-A | 440 | 48.80 | 6.24 | 48.98 | 78.91 | -0.338 | 叶绿体 | |||||
| BnA05g0211550.1 | BnLPAT9-A | 376 | 43.89 | 9.05 | 37.63 | 93.56 | 0.043 | 内质网 | |||||
| BnA05g0213610.1 | BnLPAT10-A | 531 | 60.03 | 8.06 | 42.27 | 92.34 | -0.076 | 线粒体 | |||||
| BnA07g0286470.1 | BnLPAT11-A | 399 | 44.75 | 6.13 | 53.03 | 87.42 | -0.182 | 叶绿体,内质网 | |||||
| BnA07g0288480.1 | BnLPAT12-A | 380 | 43.26 | 8.25 | 44.55 | 98.76 | 0.144 | 内质网 | |||||
| BnA07g0302330.1 | BnLPAT13-A | 287 | 31.98 | 9.30 | 47.91 | 94.11 | -0.036 | 叶绿体,内质网 | |||||
| BnA07g0303170.1 | BnLPAT14-A | 370 | 41.87 | 5.93 | 48.34 | 91.38 | -0.176 | 内质网 | |||||
| BnA08g0305940.1 | BnLPAT15-A | 345 | 39.42 | 9.14 | 34.60 | 100.06 | -0.014 | 内质网 | |||||
| BnA08g0312680.1 | BnLPAT16-A | 454 | 50.01 | 6.25 | 43.76 | 78.00 | -0.331 | 叶绿体 | |||||
| BnA08g0327830.1 | BnLPAT17-A | 814 | 92.41 | 8.00 | 49.32 | 96.38 | -0.008 | - | |||||
| BnA09g0362990.1 | BnLPAT18-A | 451 | 49.63 | 6.32 | 44.65 | 83.28 | -0.202 | 叶绿体 | |||||
| BnA09g0374960.1 | BnLPAT19-A | 390 | 43.76 | 9.38 | 47.42 | 99.51 | 0.105 | 内质网 | |||||
| BnA09g0384080.1 | BnLPAT20-A | 358 | 39.41 | 9.67 | 43.09 | 87.37 | 0.037 | 叶绿体 | |||||
| BnA10g0406100.1 | BnLPAT21-A | 326 | 37.21 | 8.70 | 47.19 | 87.30 | -0.132 | 叶绿体,内质网 | |||||
| BnC01g0428330.1 | BnLPAT1-C | 376 | 43.22 | 9.05 | 47.12 | 88.88 | -0.099 | 内质网 | |||||
| BnC01g0463380.1 | BnLPAT2-C | 339 | 39.24 | 8.67 | 42.96 | 92.27 | 0.085 | 内质网 | |||||
| BnC02g0500200.1 | BnLPAT13-C | 285 | 31.86 | 9.24 | 49.19 | 96.46 | -0.049 | 叶绿体,内质网 | |||||
| BnC03g0553040.1 | BnLPAT18-C | 385 | 42.78 | 5.86 | 46.51 | 78.29 | -0.367 | 叶绿体 | |||||
| BnC03g0566190.1 | BnLPAT3-C | 534 | 60.30 | 5.85 | 39.86 | 90.54 | -0.057 | 线粒体,细胞核 | |||||
| BnC03g0576540.1 | BnLPAT5-C | 432 | 48.17 | 9.74 | 43.64 | 93.63 | -0.214 | 叶绿体,内质网 | |||||
| BnC03g0615450.1 | BnLPAT6-C | 286 | 31.79 | 9.39 | 44.60 | 94.44 | -0.030 | 叶绿体,内质网 | |||||
| BnC03g0616110.1 | BnLPAT15-C | 333 | 37.86 | 8.40 | 31.42 | 96.31 | 0.069 | 内质网 | |||||
| BnC04g0635520.1 | BnLPAT12-C | 376 | 43.16 | 6.46 | 48.00 | 94.63 | 0.112 | 内质网 | |||||
| BnC04g0683780.1 | BnLPAT10-C | 541 | 61.22 | 6.68 | 43.10 | 90.61 | -0.089 | 线粒体,细胞核 | |||||
| BnC05g0715370.1 | BnLPAT8-C | 451 | 49.88 | 6.33 | 50.53 | 77.43 | -0.359 | 叶绿体 | |||||
| BnC06g0752240.1 | BnLPAT4-C | 391 | 43.66 | 9.48 | 45.11 | 99.77 | 0.111 | 内质网 | |||||
| BnC06g0756180.1 | BnLPAT11-C | 397 | 44.50 | 6.13 | 54.75 | 89.82 | -0.167 | 内质网 | |||||
| BnC06g0775100.1 | BnLPAT22-C | 383 | 43.04 | 5.86 | 49.72 | 88.54 | -0.161 | 内质网 | |||||
| BnC06g0775680.1 | BnLPAT14-C | 381 | 43.06 | 6.19 | 50.25 | 89.00 | -0.175 | 内质网 | |||||
| BnC07g0780780.1 | BnLPAT23-C | 492 | 55.49 | 5.99 | 40.00 | 89.15 | -0.073 | 内质网 | |||||
| BnC07g0830090.1 | BnLPAT20-C | 345 | 37.98 | 9.65 | 42.19 | 89.54 | 0.025 | 叶绿体 | |||||
| BnC08g0842420.1 | BnLPAT16-C | 456 | 50.22 | 6.14 | 44.19 | 79.58 | -0.302 | 叶绿体 | |||||
| BnC08g0866950.1 | BnLPAT19-C | 390 | 43.73 | 9.38 | 46.43 | 99.51 | 0.106 | 内质网 | |||||
| BnC09g0920060.1 | BnLPAT21-C | 371 | 42.51 | 8.92 | 47.19 | 92.18 | -0.031 | 内质网 | |||||
| BnUnng0943360.1 | BnLPAT9-U | 346 | 40.38 | 9.01 | 39.34 | 90.69 | 0.032 | 内质网 | |||||
| BnUnng0994230.1 | BnLPAT12-U | 380 | 43.26 | 8.74 | 45.79 | 96.71 | 0.116 | 细胞膜,内质网 | |||||
| BnUnng1007920.1 | BnLPAT2-U | 378 | 43.65 | 8.73 | 38.59 | 95.37 | 0.096 | 内质网 | |||||
未预测到BnLPAT17-A的亚细胞定位。
The subcellular localization of BnLPAT17-A is not predicted.
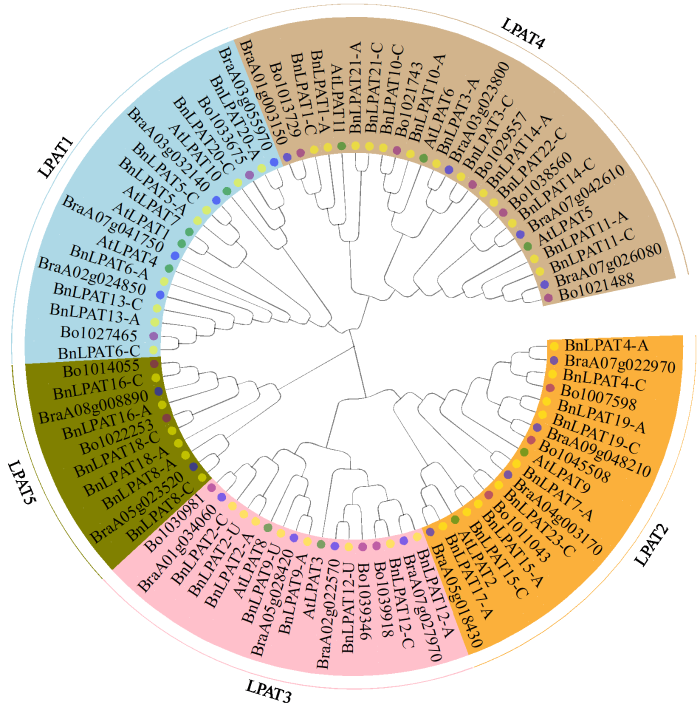
2.2 甘蓝型油菜LPAT蛋白系统发育分析
为了获取甘蓝型油菜LPAT蛋白的系统发育关系,构建了一个来自双子叶模式植物拟南芥(Arabidopsis)、白菜(Brassica rapa)、甘蓝(Brassica oleracea)和甘蓝型油菜共88个LPAT成员的临近加入法的系统发育进化树(图1),其中拟南芥有11个LPAT成员,白菜有18个,甘蓝有15个,甘蓝型油菜有44个。结果(图1)显示,88个LPAT蛋白分为5个亚家族。其中,LPAT1亚家族成员包含18个成员,包括4个拟南芥、4个白菜、2个甘蓝和8个甘蓝型油菜LPAT成员;LPAT2亚家族有18个成员,包括2个拟南芥、4个白菜、3个甘蓝以及9个甘蓝型油菜LPAT成员;LPAT3亚家族有17个成员,包括2个拟南芥、4个白菜、3个甘蓝和8个甘蓝型油菜LPAT成员;LPAT4亚家族有25个成员,数目最多,包括3个拟南芥、4个白菜、5个甘蓝和13个油菜LPAT成员;没有拟南芥LPAT成员被划到LPAT5亚家族,此分支由2个白菜、2个甘蓝和6个甘蓝型油菜LPAT成员组成。
图1
图1
拟南芥、白菜、甘蓝和甘蓝型油菜LPAT成员系统发育进化树
绿色、蓝色、紫色和黄色圆点代表分别来自拟南芥、白菜、甘蓝和甘蓝型油菜的成员。
Fig.1
Phylogenetic tree of LPAT members in Arabidopsis, B. rapa, B. oleracea and B. napus
The green, blue, purple, and yellow dots represent members from Arabidopsis, B. rapa, B. oleracea and B. napus, respectively.
白菜和甘蓝中分别有18和15个LPAT成员,总和小于甘蓝型油菜LPAT成员数量,这可能是在甘蓝型油菜在进化过程中,LPAT家族部分成员发生了基因扩增事件。例如LPAT1亚家族BnLPAT5仅在白菜中发现了同源成员Bra03g032140,而在甘蓝中没有相应的同源成员;相似地,BnLPAT6和BnLPAT13在白菜和甘蓝中仅有1对同源基因BraA02g024850和Bol027465,同时如Bol030981和BraA01g034060在甘蓝型油菜中产生了3个BnLPAT2基因的拷贝,这些结果都表明甘蓝型油菜LPAT家族基因在进化中发生了由基因复制驱动的扩张;但一些基因在甘蓝型油菜进化过程中出现与白菜、甘蓝保守相同的特点,如LPAT2亚家族的BnLPAT4 2个同源基因和BraA07g022970以及Bol007598显示较为保守的进化关系。
2.3 BnLPAT基因染色体定位和复制关系
图2
图2
BnLPAT基因染色体定位与复制关系
不同颜色代表不同的染色体,棕色线段两端的基因代表具有片段复制关系。
Fig.2
The chromosomal localization and segmental duplicated pairs of the BnLPAT genes
Different chromosomes are marked with different colors, and the genes at both ends of the brown line represent the genes that have segmental duplicated relationships.
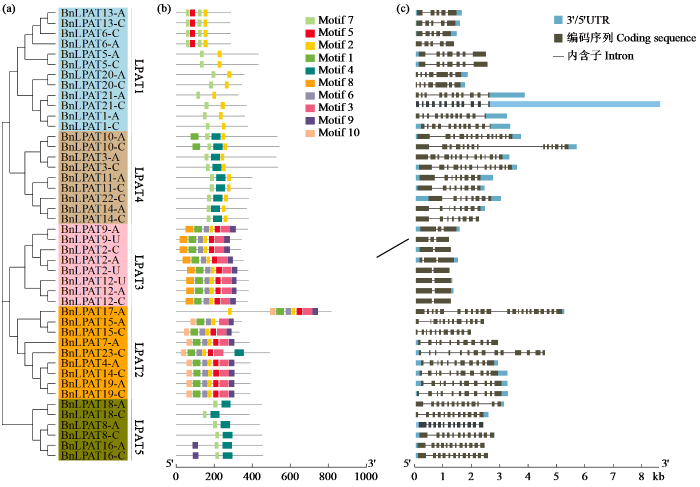
2.4 BnLPATs保守基序和基因结构分析
在BnLPATs序列上总共鉴定了10个保守Motif(图3a~b),结果显示相邻分支的成员有着保守的基序组成,如在LPAT3分支上的8个BnLPAT成员的基序组成均为Motif 1、Motif 2、Motif 3、Motif 5、Motif 6和Motif 9,此外一些基序是在特定的分支上保守出现,比如Morif 2出现在除LPAT5分支的其他分支上,Motif 3和Motif 6出现在LPAT2和LPAT3分支,Motif 8只出现在LPAT3分支上,Motif 10只出现在LPAT2分支上,这些结果暗示了不同分支的生物学功能特征可能是这些保守基序导致的。
图3
图3
BnLPATs保守基序和基因结构
(a) 44个BnLAPT临近加入法聚类树;(b) BnLPAT保守基序,共10个基序被检测,不同的基序用不同颜色的矩形表示;(c) BnLPAT基因结构,编码序列用灰色矩形表示,上、下游非编码区用蓝色矩形表示,内含子序列用线段表示。
Fig.3
Conserved motifs and gene structure of BnLPATs
(a) Neighbor-Joining clustering trees of 44 BnLPATs; (b) BnLPATs conserved motifs, with 10 motifs detected. Different motifs are represented by differently colored rectangles. (c) Gene structure of BnLPAT. The coding sequence is represented by a gray rectangle, the upstream and downstream non-coding regions are represented by a blue rectangle, and the intron sequence is represented by a line.
不同分支的BnLPAT基因结构组成相似(图3c),LPAT3分支的基因内含子数目最少,由2或3个内含子组成,其他分支的BnLPAT基因组成较为复杂,内含子数目在5(BnLPAT6-A、BnLPAT6-C、BnLPAT13-A和BnLPAT13-C)~22个(BnLPAT17-A)不等。
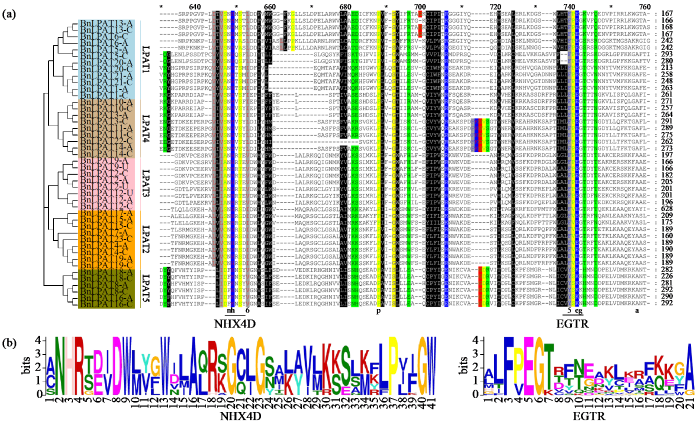
图4
图4
BnLPAT蛋白的序列比对
(a) BnLPAT蛋白的ClustalW法多序列比对;(b) 2个保守结构域的LOGO组成。
Fig.4
Sequence alignment of BnLPAT proteins
(a) ClustalW method for multiple sequence alignment of BnLPAT proteins; (b) The LOGO composition of two conserved domains.
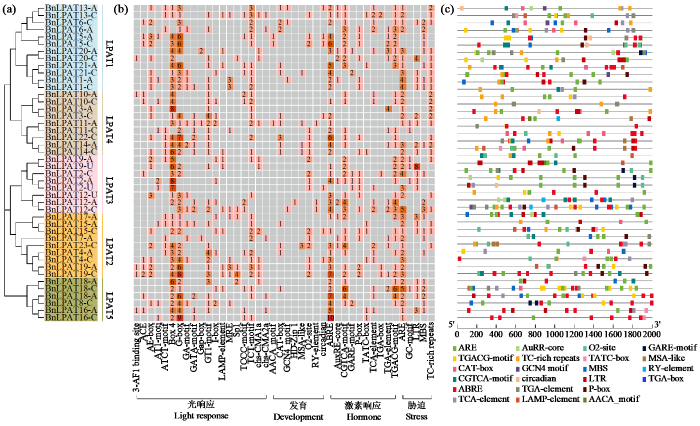
2.5 顺式作用元件分析
在PlantCARE数据库上提交BnLPAT基因上游2000 bp序列作为推定的启动子区域,总共鉴定到42种共1098个顺式作用元件,并根据其出现的位置将22种特异性响应元件可视化(图5)。按照响应类型将其分为4类,如Box 4和G-box等19种光响应元件共出现了524次;与生长发育相关的元件如CAT-box(与分生组织表达相关)和O2-site(玉米醇溶蛋白代谢);与激素响应相关的元件共10种,其中ABRE元件(参与脱落酸响应)是出现次数最多的,共114次,此外如赤霉素(GA)响应元件GARE-motif和P-box以及茉莉酸甲酯(MeJA)响应元件CGTCA-motif和TGACG-motif等也出现在BnLPAT启动子区域;与逆境胁迫响应相关的5种元件如ARE(缺氧诱导)出现在38个BnLPAT基因启动子区域,与干旱响应相关的元件MBS出现在21个BnLPAT基因启动子区域。上述结果说明了BnLPAT基因可能受不同的外界环境调控表达。
图5
图5
BnLPAT基因顺式作用元件分析
(a) 44个BnLAPT临近加入法聚类树;(b) 42种顺式作用元件按照相应类型分为4类,热图中的数字代表着元件出现的次数;(c) 22种顺式作用元件在BnLPAT基因上游2000 bp区域出现的位置,不同的顺式作用元件用不同颜色的矩形表示。
Fig.5
Analysis of BnLPAT gene cis-elements
(a) Neighbor-Joining clustering trees of 44 BnLPATs; (b) 42 cis-elements are classified into four types according to their corresponding types, and the numbers in the heat map represent the number of occurrences of the elements; (c) The positions of 22 cis-elements in the 2000 bp upstream region of the BnLPAT genes, with different cis-elements represented by different colored rectangles.
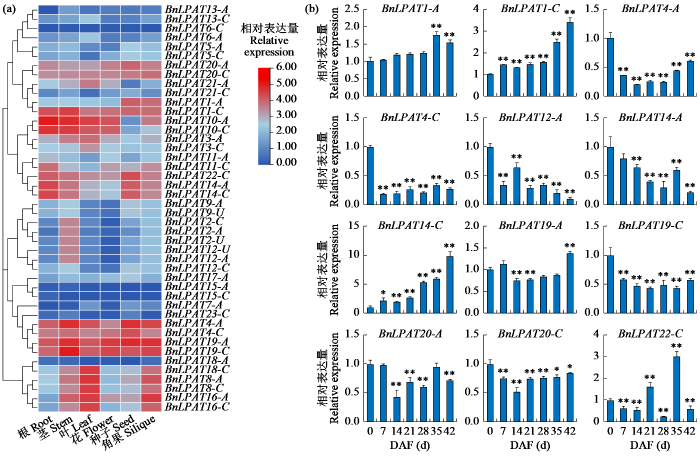
2.6 BnLPAT基因组织表达模式与种子发育过程中表达分析
根据BnTIR数据库上ZS11转录组数据,绘制BnLPAT基因表达谱热图(图6a),44个基因中除BnLPAT6-C、BnLPAT15-A和BnLPAT15-C外的41个基因在6个组织中均有表达。其中BnLPAT1-C、BnLPAT4-A、BnLPAT4-C、BnLPAT19-A、BnLPAT19- C、BnLPAT20-A、BnLPAT20-C和BnLPAT22-C在根、茎、叶、花、种子和角果中均有较高的表达量,BnLPAT1-A、BnLPAT14-A和BnLPAT14-C在种子中有较高的表达,BnLPAT10-A和BnLPAT10-C在根和茎中高表达,BnLPAT16-A和BnLPAT16-C在叶和角果中高表达。
图6
图6
BnLPAT基因表达模式
(a) 不同组织部位的BnLPAT基因转录组表达谱;(b) 12个BnLPAT基因在种子发育过程中的表达模式,*:0.01 < P < 0.05;**:P < 0.01。下同。
Fig.6
Expression pattern of BnLPAT genes
(a) Transcriptome expression profile of BnLPAT genes in different tissues. (b) Expression pattern of 12 BnLPAT genes during seed development, *: 0.01 < P < 0.05; **: P < 0.01. The same below.
研究[25-26]表明LPAT成员在植物脂肪酸积累方面发挥着重要作用,甘蓝型油菜是以种子为收获目的的油料作物,因此我们根据表达谱对ZS11种子发育过程中7个时期的11个种子高表达基因以及种子低表达基因BnLPAT12-A进行了qRT-PCR试验(图6b)。4个基因(BnLPAT1-A、BnLPAT1-C、BnLPAT14-C和BnLPAT19-A)整体上随着种子发育的完全,其表达量呈现上升趋势,3个基因(BnLPAT12-C、BnLPAT14-A和BnLPAT19-C)的表达随着种子发育呈现下降趋势,此外BnLPAT4-A和BnLPAT20-C的表达先下降后上升,BnLPAT4-C、BnLPAT19-C和BnLPAT20-C在花后0 d表达量最高,BnLPAT22-C在花后35 d表达量最高。
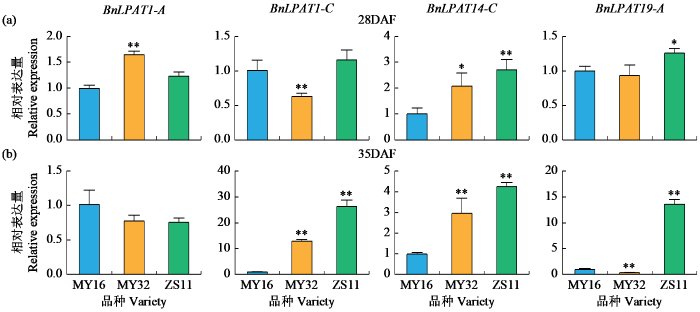
进一步于花后28和35 d在3个含油量不同的材料中检测了BnLPAT1-A、BnLPAT1-C、BnLPAT14-C和BnLPAT20-C这4个在种子发育过程中上调的基因表达特点。结果如图7所示,在花后28和35 d时,BnLPAT14-C在含油量高的品种ZS11中的表达量显著高于含油量中、低的品种;BnLPAT1-C在花后35 d时在ZS11中的表达量高于其他2个品种;BnLPAT19-A在28 d时在3个品种中表达量差距不大,但在35 d时在ZS11中的表达量显著高于其他2个品种。除BnLPAT1-A外的3个基因在花后28或35 d均表现出在高油材料背景下表达上调的特点,可见这3个基因参与了油菜种子脂肪酸的积累。
图7
图7
4个BnLPAT基因在不同品种的表达模式
Fig.7
Expression pattern of four BnLPAT genes in different varieties
3 讨论
研究脂肪酸合成与代谢相关的基因一直是油料作物的热点,研究[9]表明LPAT家族成员在脂肪酸的积累方面起到调控作用,如在拟南芥中过表达甘蓝型油菜BAT1.13(本研究中的BnLPAT19-C)和BAT1.5(BnLPAT4-C)提高了转基因拟南芥种子含油量,而这2个基因在种子中高表达。在拟南芥中过表达酵母LPAT基因SLC1提高了转基因株系的含油量[27],在烟草中过表达紫苏PfLPAT2提高了叶片的含油量[28]。截至目前,已经在许多植物全基因组水平上鉴定到LPAT成员,但在油料作物甘蓝型油菜中尚未有系统性的研究。在本研究中,在全基因组水平上共鉴定到了44个BnLPAT成员,并分析基因结构、保守基序、顺式作用元件和组织表达模式以及种子发育过程中的表达。
按照系统发育关系,44个BnLPAT被分为5个分支,其中LPAT5分支的6个BnLPATs较AtLPATs同源关系较远,同时基因结构、保守基序也暗示了这些BnLPAT蛋白可能具有不同于拟南芥LPAT成员的生物学功能,其余4个分支的LPAT在进化中保守。有研究[29]表明,质体型AtLPAT1(本研究中命名为AtLPAT10)与胚胎发育相关,AtLPAT1定位在叶绿体中,atlpat1突变体出现胚胎致死的现象,BnLPAT20的2个同源基因和AtLPAT1有着较近系统发育关系,并且BnLPAT20-A和BnLPAT20-C在种子高表达,因此推断这2个基因可能也在胚胎发育中发挥功能。
基因家族是由某些古基因经过复制产生2个或者以上的拷贝而构成的一组基因[30],基因复制事件大部分由串联复制和片段复制组成[31]。构成甘蓝型油菜BnLPAT基因复制事件均发生在染色体不同区域而被认定为片段复制[32],这可能是虽然甘蓝型油菜基因组远小于棉花基因组,但LPAT成员数目仍然大于棉花LPAT成员数目,也是导致BnLPAT数目大于白菜和甘蓝LPAT数目的原因,如在LPAT4分支中,BnLPAT基因数目分别是白菜和甘蓝LPAT基因数目的3.25和2.60倍。基因结构、保守基序和序列比对结果表明,相邻分支的BnLPAT成员在进化中较为保守,这些结果暗示了这些基因可能具有相似的生物学功能。先前的研究[11,33]表明,除调控脂质物质的合成代谢外,LPAT还在育性、抗逆性以及膜重构方面发挥作用,例如在低温环境中拟南芥fab1突变体不能维持正常的光合作用,这是由于fab1突变体在低温下高熔点的磷脂酰甘油含量以及膜脂中16:0脂肪酸含量增加导致低温损伤[34];在白念珠菌(Candida albicas)中过表达LPT1基因,当提供棕榈酸时,膜磷脂表现为不均匀分布[35];此外过表达LPAAT3通过改变磷脂和溶血磷脂抑制了高尔基体膜小管的形成[36]。大量顺式作用元件被鉴定到存在BnLPAT基因启动子区域,比如在LPAT5分支上的成员BnLPAT16-C存在10个ABRE元件,BnLPAT9-U存在8个LTR元件,暗示这2个基因表达可能分别受脱落酸以及低温胁迫的调控,从而在不同环境下发挥生物学功能。
基因的表达模式与其功能密切相关,转录组表达分析表明,不同分支的BnLPAT具有特异性的表达特点,例如LPAT1分支多数成员表达不高,LPAT2分支的4个成员在不同的组织均有较高的表达量,LPAT3分支的成员在茎中有着相对较高的表达,LPAT4分支成员在根、茎、叶和种子有相对较高的表达量,而LPAT5成员在叶和角果中表达最高。植物根部往往最先响应胁迫信号,根特异表达的基因通常和逆境适应相关,因此LPAT4分支的成员可能在抗逆适应发挥功能。11个在种子高表达的基因,有4个(BnLPAT1-A、BnLPAT1-C、BnLPAT14- C和BnLPAT19-A)随着种子的发育其表达量逐渐升高。有研究[37]表明,油菜种子中脂肪酸的积累也是随着时间的增加而增加,在35DAF左右脂肪酸含量达到最大,随后脂肪酸合成达到稳定。因此,我们在3个含油量不同的材料中检测了这4个基因的表达量,其中3个基因在28或者35 DAF时,高含油量的品种基因表达水平显著高于其他2个品种的表达水平。这些结果暗示了这3个基因可能在油菜种子脂肪酸积累方面发挥功能。总之,甘蓝型油菜LPAT基因家族表达模式分析能为进一步研究其生物学功能提供线索。
4 结论
甘蓝型油菜高质量基因组的组装发布以及高效遗传转化体系的建立为油菜分子育种提供了可能,而利用转基因或者基因编辑手段对调控油脂合成通路基因进行干预从而获得高品质油菜种质方兴未艾,但是在全基因组水平对油菜脂肪酸合成关键基因家族LAPT的鉴定尚未见报道。此项研究在鉴定BnLPAT家族成员的基础上进行了分类,并开展了系统的生物信息学相关分析,同时利用转录组数据以及qRT-PCR技术研究了BnLPAT基因的表达模式,初步明确了在脂肪酸合成过程的关键基因。
参考文献
Regulation of lipid synthesis in oil crops
DOI:10.1016/j.febslet.2013.05.018
PMID:23684640
[本文引用: 1]

Oil crops are in increasing demand both for food and as renewable sources of chemicals. It is therefore vital to understand how oil accumulation is regulated. Different ways of obtaining such information are discussed with an emphasis on metabolic control analysis. The usefulness of the latter has been well-illustrated by its application to help raise yields in oilseed rape.Copyright © 2013. Published by Elsevier B.V.
The Kennedy pathway—De novo synthesis of phosphatidylethanolamine and phosphatidylcholine
DOI:10.1002/iub.337
PMID:20503434
[本文引用: 1]

The glycerophospholipids phosphatidylcholine (PC) and phosphatidylethanolamine (PE) account for greater than 50% of the total phospholipid species in eukaryotic membranes and thus play major roles in the structure and function of those membranes. In most eukaryotic cells, PC and PE are synthesized by an aminoalcoholphosphotransferase reaction, which uses sn-1,2-diradylglycerol and either CDP-choline or CDP-ethanolamine, respectively. This is the last step in a biosynthetic pathway known as the Kennedy pathway, so named after Eugene Kennedy who elucidated it over 50 years ago. This review will cover various aspects of the Kennedy pathway including: each of the biosynthetic steps, the functions and roles of the phospholipid products PC and PE, and how the Kennedy pathway has the potential of being a chemotherapeutic target against cancer and various infectious diseases.
Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues
Molecular evolution of the lysophosphatidic acid acyltransferase (LPAAT) gene family
The structure and functions of human lysophosphatidic acid acyltransferases
Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis
DOI:10.1104/pp.109.148247
PMID:19965969
[本文引用: 2]

In higher plants, lysophosphatidic acid acyltransferase (LPAAT), located in the cytoplasmic endomembrane compartment, plays an essential role in the synthesis of phosphatidic acid, a key intermediate in the biosynthesis of membrane phospholipids in all tissues and storage lipids in developing seeds. In order to assess the contribution of LPAATs to the synthesis of storage lipids, we have characterized two microsomal LPAAT isozymes, the products of homoeologous genes that are expressed in rapeseed (Brassica napus). DNA sequence homologies, complementation of a bacterial LPAAT-deficient mutant, and enzymatic properties confirmed that each of two cDNAs isolated from a Brassica napus immature embryo library encoded a functional LPAAT possessing the properties of a eukaryotic pathway enzyme. Analyses in planta revealed differences in the expression of the two genes, one of which was detected in all rapeseed tissues and during silique and seed development, whereas the expression of the second gene was restricted predominantly to siliques and developing seeds. Expression of each rapeseed LPAAT isozyme in Arabidopsis (Arabidopsis thaliana) resulted in the production of seeds characterized by a greater lipid content and seed mass. These results support the hypothesis that increasing the expression of glycerolipid acyltransferases in seeds leads to a greater flux of intermediates through the Kennedy pathway and results in enhanced triacylglycerol accumulation.
三角褐指藻LPAAT基因在酵母中表达对脂肪酸的影响
从三角褐指藻(Phaeodactylum tricornutum)中克隆到1 632bp的溶血磷脂酸酰基转移酶(LPAAT)全长cDNA,其中包含的开放阅读框被命名为PtLPAAT。在毕赤酵母中异源表达PtLPAAT基因,诱导培养45h时随机选取的3个酵母转化子总脂肪酸含量分别提高了2.68%、11.14%、31.28%;诱导培养72h时同样的转化子总脂肪酸含量分别提高11.59%、18.72%、46.63%。用薄层层析法分离48h后的酵母菌体的甘油三酯,并用气相色谱法测定其脂肪酸,结果显示:与非转基因对照菌株相比,其油酸提高了0.9倍、亚油酸提高了5倍。用荧光定量PCR检测发现PtLPAAT基因的表达量随三角褐指藻油脂不断积累逐渐升高。表明在酵母中表达PtLPAAT基因不仅能够明显地提高其脂肪酸含量,而且还能选择性结合不饱和脂肪酸到甘油三酯中。
Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis
A genome-wide analysis of the lysophosphatidate acyltransferase (LPAAT) gene family in cotton: organization, expression, sequence variation, and association with seed oil content and fiber quality
DOI:10.1186/s12864-017-3594-9
PMID:28249560
[本文引用: 1]

Background: Lysophosphatidic acid acyltransferase (LPAAT) encoded by a multigene family is a rate-limiting enzyme in the Kennedy pathway in higher plants. Cotton is the most important natural fiber crop and one of the most important oilseed crops. However, little is known on genes coding for LPAATs involved in oil biosynthesis with regard to its genome organization, diversity, expression, natural genetic variation, and association with fiber development and oil content in cotton.Results: In this study, a comprehensive genome-wide analysis in four Gossypium species with genome sequences, i. e., tetraploid G. hirsutum-AD(1) and G. barbadense-AD(2) and its possible ancestral diploids G. raimondii-D-5 and G. arboreumA(2), identified 13, 10, 8, and 9 LPAAT genes, respectively, that were divided into four subfamilies. RNA-seq analyses of the LPAAT genes in the widely grown G. hirsutum suggest their differential expression at the transcriptional level in developing cottonseeds and fibers. Although 10 LPAAT genes were co-localised with quantitative trait loci (QTL) for cottonseed oil or protein content within a 25-cM region, only one single strand conformation polymorphic (SSCP) marker developed from a synonymous single nucleotide polymorphism (SNP) of the At-Gh13LPAAT5 gene was significantly correlated with cottonseed oil and protein contents in one of the three field tests. Moreover, transformed yeasts using the At-Gh13LPAAT5 gene with the two sequences for the SNP led to similar results, i. e., a 25-31% increase in palmitic acid and oleic acid, and a 16-29% increase in total triacylglycerol (TAG).Conclusions: The results in this study demonstrated that the natural variation in the LPAAT genes to improving cottonseed oil content and fiber quality is limited; therefore, traditional cross breeding should not expect much progress in improving cottonseed oil content or fiber quality through a marker-assisted selection for the LPAAT genes. However, enhancing the expression of one of the LPAAT genes such as At-Gh13LPAAT5 can significantly increase the production of total TAG and other fatty acids, providing an incentive for further studies into the use of LPAAT genes to increase cottonseed oil content through biotechnology.
Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus
Application of developmental regulators to improve in planta or in vitro transformation in plants
Clustal W and Clustal X version 2.0
DOI:10.1093/bioinformatics/btm404
PMID:17846036
[本文引用: 1]

The Clustal W and Clustal X multiple sequence alignment programs have been completely rewritten in C++. This will facilitate the further development of the alignment algorithms in the future and has allowed proper porting of the programs to the latest versions of Linux, Macintosh and Windows operating systems.The programs can be run on-line from the EBI web server: http://www.ebi.ac.uk/tools/clustalw2. The source code and executables for Windows, Linux and Macintosh computers are available from the EBI ftp site ftp://ftp.ebi.ac.uk/pub/software/clustalw2/
MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets
DOI:10.1093/molbev/msw054
PMID:27004904
[本文引用: 1]

We present the latest version of the Molecular Evolutionary Genetics Analysis (Mega) software, which contains many sophisticated methods and tools for phylogenomics and phylomedicine. In this major upgrade, Mega has been optimized for use on 64-bit computing systems for analyzing larger datasets. Researchers can now explore and analyze tens of thousands of sequences in Mega The new version also provides an advanced wizard for building timetrees and includes a new functionality to automatically predict gene duplication events in gene family trees. The 64-bit Mega is made available in two interfaces: graphical and command line. The graphical user interface (GUI) is a native Microsoft Windows application that can also be used on Mac OS X. The command line Mega is available as native applications for Windows, Linux, and Mac OS X. They are intended for use in high-throughput and scripted analysis. Both versions are available from www.megasoftware.net free of charge.© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Extent of gene duplication in the genomes of Drosophila, nematode, and yeast
TBtools: an integrative toolkit developed for interactive analyses of big biological data
DOI:S1674-2052(20)30187-8
PMID:32585190
[本文引用: 1]

The rapid development of high-throughput sequencing techniques has led biology into the big-data era. Data analyses using various bioinformatics tools rely on programming and command-line environments, which are challenging and time-consuming for most wet-lab biologists. Here, we present TBtools (a Toolkit for Biologists integrating various biological data-handling tools), a stand-alone software with a user-friendly interface. The toolkit incorporates over 130 functions, which are designed to meet the increasing demand for big-data analyses, ranging from bulk sequence processing to interactive data visualization. A wide variety of graphs can be prepared in TBtools using a new plotting engine ("JIGplot") developed to maximize their interactive ability; this engine allows quick point-and-click modification of almost every graphic feature. TBtools is platform-independent software that can be run under all operating systems with Java Runtime Environment 1.6 or newer. It is freely available to non-commercial users at https://github.com/CJ-Chen/TBtools/releases.Copyright © 2020 The Author. Published by Elsevier Inc. All rights reserved.
Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method
DOI:10.1006/meth.2001.1262
PMID:11846609
[本文引用: 1]

The two most commonly used methods to analyze data from real-time, quantitative PCR experiments are absolute quantification and relative quantification. Absolute quantification determines the input copy number, usually by relating the PCR signal to a standard curve. Relative quantification relates the PCR signal of the target transcript in a treatment group to that of another sample such as an untreated control. The 2(-Delta Delta C(T)) method is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments. The purpose of this report is to present the derivation, assumptions, and applications of the 2(-Delta Delta C(T)) method. In addition, we present the derivation and applications of two variations of the 2(-Delta Delta C(T)) method that may be useful in the analysis of real-time, quantitative PCR data.Copyright 2001 Elsevier Science (USA).
Application of student's t-test, analysis of variance, and covariance
Duplication and divergence: the evolution of new genes and old ideas
Functional characterization of human 1-acylglycerol-3-phosphate-O- acyltransferase isoform 9: cloning, tissue distribution, gene structure, and enzymatic activity
A review of erucic acid production in brassicaceae oilseeds: progress and prospects for the genetic engineering of high and low-erucic acid rapeseeds (Brassica napus)
The lysophosphatidic acid acyltransferases (acylglycerophosphate acyltransferases) family: one reaction, five enzymes, many roles
DOI:10.1097/MOL.0000000000000492
PMID:29373329
[本文引用: 1]

Lysophosphatidic acid acyltransferases (LPAATs)/acylglycerophosphate acyltransferases (AGPATs) are a homologous group of enzymes that all catalyze the de novo formation of phosphatidic acid from lysophosphatidic acid (LPA) and a fatty acyl-CoA. This review seeks to resolve the apparent redundancy of LPAATs through examination of recent literature.Recent molecular studies suggest that individual LPAAT homologues produce functionally distinct pools of phosphatidic acid, whereas gene ablation studies demonstrate unique roles despite a similar biochemical function. Loss of the individual enzymes not only causes diverse effects on down-stream lipid metabolism, which can vary even for a single enzyme from one tissue to the next, but also results in a wide array of physiological consequences, ranging from cognitive impairment, to lipodystrophy, to embryonic lethality.LPAATs are critical mediators of cell membrane phospholipid synthesis, regulating the production of specific down-stream glycerophospholipid species through generation of distinct pools of phosphatidic acid that feed into dedicated biosynthetic pathways. Loss of any specific LPAAT can lead to alterations in cellular and organellar membrane phospholipid composition that can vary for a single enzyme in different tissues, with unique pathophysiological implications.
Changes in oil content of transgenic soybeans expressing the yeast SLC1 gene
Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis
Phosphatidic acid is a key intermediate for chloroplast membrane lipid biosynthesis. De novo phosphatidic acid biosynthesis in plants occurs in two steps: first the acylation of the sn-1 position of glycerol-3-phosphate giving rise to lysophosphatidic acid; second, the acylation of the sn-2 position of lysophosphatidic acid to form phosphatidic acid. The second step is catalyzed by a lysophosphatidic acid acyltransferase (LPAAT). Here we describe the identification of the ATS2 gene of Arabidopsis encoding the plastidic isoform of this enzyme. Introduction of the ATS2 cDNA into E. coli JC 201, which is temperature-sensitive and carries a mutation in its LPAAT gene plsC, restored this mutant to nearly wild type growth at high temperature. A green-fluorescent protein fusion with ATS2 localized to the chloroplast. Disruption of the ATS2 gene of Arabidopsis by T-DNA insertion caused embryo lethality. The development of the embryos was arrested at the globular stage concomitant with a transient increase in ATS2 gene expression. Apparently, plastidic LPAAT is essential for embryo development in Arabidopsis during the transition from the globular to the heart stage when chloroplasts begin to form.
The early stages of duplicate gene evolution
Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth
The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana
Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis
A mutation in the LPAT1 gene suppresses the sensitivity of fab 1 plants to low temperature
Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity
Lysophosphatidic acid acyltransferase 3 regulates Golgi complex structure and function