作物杂志,2018, 第6期: 58–67 doi: 10.16035/j.issn.1001-7283.2018.06.010
小麦GASA基因家族生物信息学分析
吕亮杰,陈希勇,张业伦,刘茜,王莉梅,马乐,李辉
- 河北省农林科学院粮油作物研究所/河北省作物遗传育种实验室,050035,河北石家庄
Bioinformatics Identification of GASA Gene Family Expression Profiles in Wheat
Lü Liangjie,Chen Xiyong,Zhang Yelun,Liu Qian,Wang Limei,Ma Le,Li Hui
- Hebei Academy of Agriculture and Forestry Sciences, Institute of Cereal and Oil Crops/Crop Genetics and Breeding Laboratory of Hebei, Shijiazhuang 050035, Hebei, China
摘要:
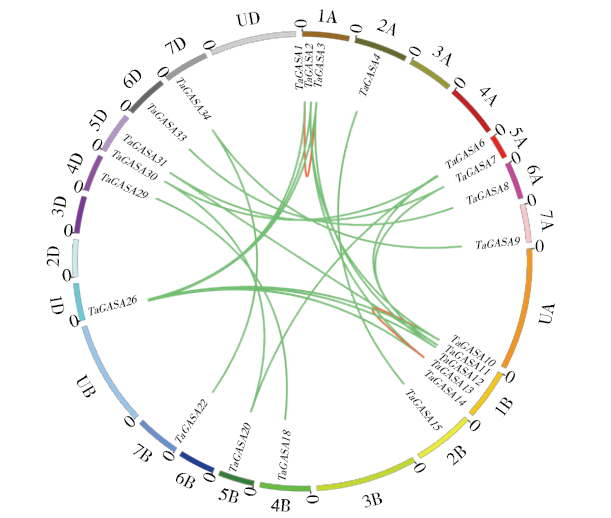
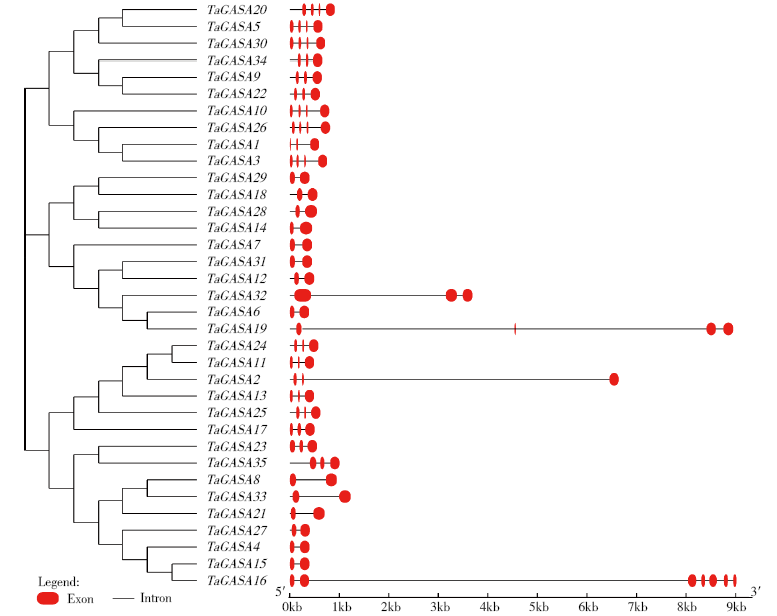
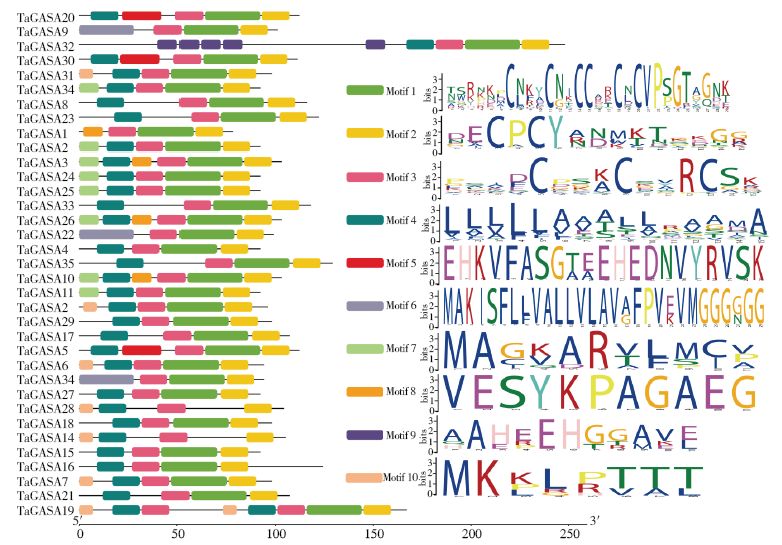
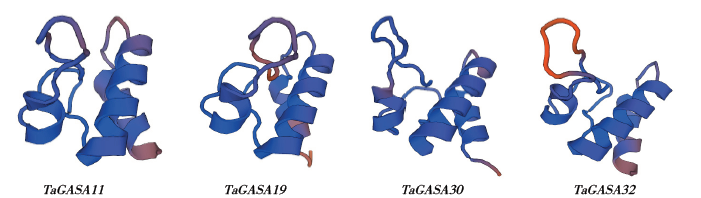
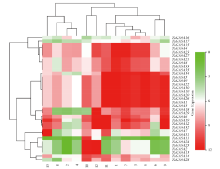
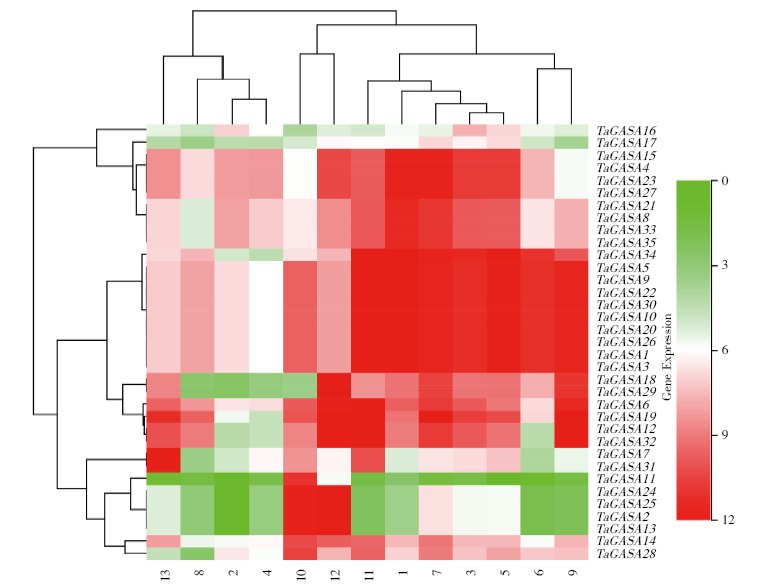
赤霉素调节的GASA(Gibberellic Acid-Stimulated in Arabidopsis)基因家族是植物特有的转录因子家族,在调控植物生长发育过程中起重要作用。关于小麦GASA基因家族全基因组分析的研究尚未见报道。为进一步探讨小麦GASA基因的功能,通过对小麦最新基因组数据进行分析,获得了35个TaGASA基因,命名为TaGASAs,根据染色体编号排列为TaGASA1~TaGASA35。结合公布的小麦品种Chinese Spring的基因组数据,采用生物信息学方法对其基因结构、染色体分布、蛋白结构、系统进化及表达谱进行了分析。结果表明,35个小麦TaGASA基因分布于除3A、4A、3B、3D染色体外的其余17条染色体上,基因编码长度为78~264个氨基酸的蛋白质,基因外显子数量从2个到7个不等;串联重复片段分析结果表明,片段复制和串联重复是小麦TaGASA家族基因扩张的主要模式;小麦TaGASA蛋白进化树和7种作物GASA基因的系统进化树表明,GASA基因分为4个类别,同一类之间的结构较为相似;小麦35个TaGASA基因家族成员含有10个motif,推测小麦TaGASA基因家族应都含有motif1、motif2和motif3。在13个组织器官中都检测到了35个TaGASA基因的转录本,不同组织器官中TaGASA基因的表达存在明显差异。
| [1] |
Roxrud I, Lid S E, Fletcher J C , et al. GASA4,one of the 14-member Arabidopsis GASA family of small polypeptides,regulates flowering and seed development. Plant and Cell Physiology, 2007,48(3):471-483.
doi: 10.1093/pcp/pcm016 pmid: 17284469 |
| [2] |
Aubert D, Chevillard M, Dorne A M , et al. Expression patterns of GASA genes in Arabidopsis thaliana:the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Molecular Biology, 1998,36(6):871-883.
doi: 10.1023/A:1005938624418 pmid: 9520278 |
| [3] |
Wigoda N, Ben-Nissan G, Granot D , et al. The gibberellin-induced,cysteine-rich protein GIP2 from Petunia hybrida exhibits in planta antioxidant activity. Plant Journal, 2010,48(5):796-805.
doi: 10.1111/j.1365-313X.2006.02917.x pmid: 17076804 |
| [4] |
Wang L, Wang Z, Xu Y Y , et al. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant Journal, 2010,57(3):498-510.
doi: 10.1111/j.1365-313X.2008.03707.x pmid: 18980660 |
| [5] | Zhang S C, Wang X J . Expression pattern of GASA,downstream genes of DELLA,in Arabidopsis. Chinese Science Bulletin, 2008,53(24):3839-3846. |
| [6] |
Sun S L, Wang H X, Yu H M , et al. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. Journal of Experimental Botany, 2013,64(6):1637-1647.
doi: 10.1093/jxb/ert021 pmid: 23378382 |
| [7] |
Shi L F, Gast R T, Gopalraj M , et al. Characterization of a shoot-specific,GA3- and ABA-regulated gene from tomato. Plant Journal, 2010,2(2):153-159.
doi: 10.1111/j.1365-313X.1992.00153.x pmid: 1302047 |
| [8] |
Taylor B H, Scheuring C F . A molecular marker for lateral root initiation:The RSI-1 gene of tomato (Lycopersicon esculentum Mill) is activated in early lateral root primordia. Molecular & General Genetics, 1994,243(2):148-157.
doi: 10.1007/BF00280311 pmid: 8177211 |
| [9] |
Ben-Nissan G, Lee J Y, Borohov A , et al. GIP,a Petunia hybrida GA-induced cysteine-rich protein:a possible role in shoot elongation and transition to flowering. Plant Journal for Cell & Molecular Biology, 2010,37(2):229-238.
doi: 10.1046/j.1365-313X.2003.01950.x pmid: 14690507 |
| [10] |
Berrocallobo M, Segura A, Moreno M , et al. Snakin-2,an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiology, 2002,128(3):951-961.
doi: 10.1104/pp.010685 |
| [11] |
Furukawa T, Sakaguchi N, Shimada H . Two OsGASR genes,rice GAST homologue genes that are abundant in proliferating tissues,show different expression patterns in developing panicles. Genes & Genetic Systems, 2006,81(3):171-180.
doi: 10.1266/ggs.81.171 pmid: 16905871 |
| [12] |
刘秋华, 罗曼, 彭建宗 , 等. 水稻OsGASR4基因及其启动子的克隆与表达分析. 华南师范大学学报, 2015(1):81-86.
doi: 10.6054/j.jscnun.2014.11.006 |
| [13] |
Kotilainen M, Helariutta Y, Mehto M , et al. GEG participates in the regulation of cell and organ shape during corolla and carpel development in Gerbera hybrida. Plant Cell, 1999,11(6):1093-1104.
doi: 10.1105/tpc.11.6.1093 |
| [14] |
De la Fuente J I AI, Castillejo C, Sanchez-Sevilla JF , et al. The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. Journal of Experimental Botany, 2006,57(19):2401-2411.
doi: 10.1093/jxb/erj213 pmid: 16804055 |
| [15] |
Liu Z H, Zhu L, Shi H Y , et al. Cotton GASL genes encoding putative gibberellin-regulated proteins are involved in response to GA signaling in fiber development. Molecular Biology Reports, 2013,40(7):4561-4570.
doi: 10.1007/s11033-013-2543-1 pmid: 23645033 |
| [16] |
张盛春, 王小菁 . 拟南芥DELLA下游的GASA基因表达研究. 科学通报, 2008,53(22):2760.
doi: 10.1360/csb2008-53-22-2760 |
| [17] |
Rubinovich L, Weiss D . The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant Journal, 2010,64(6):1018-1027.
doi: 10.1111/j.1365-313X.2010.04390.x pmid: 21143681 |
| [18] |
Zimmermann R, Sakai H, Hochholdinger F . The gibberellic acid stimulated-like gene family in maize and its role in lateral root development. Plant Physiology, 2010,152(1):356-365.
doi: 10.1104/pp.109.149054 pmid: 19926801 |
| [19] |
Zhang S C, Yang C W, Peng J Z , et al. GASA5,a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Molecular Biology, 2009,69(6):745-759.
doi: 10.1007/s11103-009-9452-7 pmid: 202020522020202020202020202020 |
| [20] |
Moyano-Cañete E, Bellido M L, García-Caparrós N , et al. FaGAST2,a strawberry ripening-related gene,acts together with FaGAST1 to determine cell size of the fruit receptacle. Plant & Cell Physiology, 2013,54(2):218-236.
doi: 10.1093/pcp/pcs167 pmid: 23231876 |
| [21] |
Blancoportales R, Lópezraéz J A, Bellido M L , et al. A strawberry fruit-specific and ripening-related gene codes for a HyPRP protein involved in polyphenol anchoring. Plant Molecular Biology, 2004,55(6):763-780.
doi: 10.1007/s11103-004-1966-4 pmid: 15604715 |
| [22] |
Mao Z C, Zheng J Y, Wang Y S , et al. The new CaSn gene belonging to the snakin family induces resistance against root-knot nematode infection in pepper. Phytoparasitica, 2011,39(2):151-164.
doi: 10.1007/s12600-011-0149-5 |
| [23] |
Zhang S C, Wang X J . Over expression of GASA5 increases the sensitivity of Arabidopsis to heat stress. Journal of Plant Physiology, 2011,168(17):2093-2101.
doi: 10.1016/j.jplph.2011.06.010 |
| [24] |
Rubinovich L, Ruthstein S, Weiss D . The Arabidopsis cysteine-rich GASA5 is a redox-active metalloprotein that suppresses gibberellin responses. Molecular Plant, 2014,7(1):244-247.
doi: 10.1093/mp/sst141 pmid: 24157610 |
| [25] |
Huang X H, Zhao Y, Wei X H , et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nature Genetics, 2011,44(1):32-39.
doi: 10.1038/ng.1018 pmid: 22138690 |
| [26] | Ling H Q, Zhao S C, Liu D C , et al. The draft genome of Triticum urartu. Nature, 2013,496:487-490. |
| [27] |
Zhang D D, Wang B N, Zhao J M , et al. Divergence in homoeolog expression of the grain length-associated gene GASR7 during wheat allohexaploidization. The Crop Journal, 2015,3(1):1-9.
doi: 10.1016/j.cj.2014.08.005 |
| [28] |
Avni R, Nave M, Barad O , et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science, 2017,357(6346):93.
doi: 10.1126/science.aan0032 pmid: 28684525 |
| [29] |
Jia J Z, Zhao S C, Kong X Y , et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature, 2013,496(7443):91-95.
doi: 10.1038/nature12028 pmid: 23535592 |
| [30] |
Ling H Q, Zhao S C, Liu D C , et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Science Foundation in China, 2013,496(2):87-90.
doi: 10.1038/nature11997 pmid: 20 |
| [31] |
Choulet F, Alberti A, Theil S , et al. Structural and functional partitioning of bread wheat chromosome 3B. Science, 2014,345(6194):1249721.
doi: 10.1126/science.1249721 pmid: 25035497 |
| [32] |
Edgar R C . MUSCLE:multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 2004,32(5):1792-1797.
doi: 10.1093/nar/gkh340 |
| [33] | Kumar S, Stecher G, Tamura K . MEGA7:Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology & Evolution, 2016,33(7):1870-1874. |
| [34] |
赵腾, 夏新莉, 尹伟伦 . 黑杨GASA基因的克隆和功能分析. 广东农业科学, 2012,39(8):138-140.
doi: 10.3969/j.issn.1004-874X.2012.08.043 |
| [35] |
李昆仑, 柏锡, 卢姗 , 等. 碱胁迫应答GsGASA1及GsGASA2基因表达特性研究. 东北农业大学学报, 2012,43(1):143-148.
doi: 10.3969/j.issn.1005-9369.2012.01.025 |
| [1] | 龙素霞,李芳芳,石书亚,赵颖佳,肖凯. 氮磷钾配施对小麦植株养分吸收利用和产量的影响[J]. 作物杂志, 2018, (6): 96–102 |
| [2] | 时丽冉,白丽荣,吕亚慈,赵明辉,赵凤梧,李会敏. 小麦杂交品种衡9966苗期耐盐性分析[J]. 作物杂志, 2018, (6): 149–153 |
| [3] | 王汉霞,单福华,田立平,马巧云,赵昌平,张风廷. 北部冬麦区冬小麦区试品种(系)的稳定性和适应性分析[J]. 作物杂志, 2018, (5): 40–44 |
| [4] | 安霞,张海军,蒋方山,吕连杰,陈军. 播期播量对不同穗型冬小麦群体及子粒产量的影响[J]. 作物杂志, 2018, (5): 132–136 |
| [5] | 王嘉楠,李小艳,魏石美,赵会杰,赵明奇,汪月霞. 5-ALA对干旱胁迫下小麦幼苗光合作用及D1蛋白的调节作用[J]. 作物杂志, 2018, (5): 121–126 |
| [6] | 杨飞,马文礼,陈永伟,张战胜,王昊. 匀播、滴灌对春小麦幼穗分化进程及产量的影响[J]. 作物杂志, 2018, (4): 84–88 |
| [7] | 罗海斌, 蒋胜理, 黄诚梅, 曹辉庆, 邓智年, 吴凯朝, 徐林, 陆珍, 魏源文. 甘蔗ScHAK10基因克隆及表达分析[J]. 作物杂志, 2018, (4): 53–61 |
| [8] | 温辉芹,程天灵,裴自友,李雪,张立生,朱玫. 山西省近年审定小麦品种的综合性状分析[J]. 作物杂志, 2018, (4): 32–36 |
| [9] | 陆梅,孙敏,任爱霞,雷妙妙,薛玲珠,高志强. 喷施叶面肥对旱地小麦生长的影响及与产量的关系[J]. 作物杂志, 2018, (4): 121–125 |
| [10] | 赵广才,常旭虹,王德梅,陶志强,王艳杰,杨玉双,朱英杰. 小麦生产概况及其发展[J]. 作物杂志, 2018, (4): 1–7 |
| [11] | 王拯,陈兆波,张胜全,任立平,高新欢,叶志杰,张风廷. BS系列杂交小麦在陕西渭南地区制种可行性分析[J]. 作物杂志, 2018, (3): 174–179 |
| [12] | 王君婵,高致富,李东升,朱冬梅,吴宏亚. 农业信息技术在小麦育种中的应用研究[J]. 作物杂志, 2018, (3): 37–43 |
| [13] | 张彬,李金秀,王震,冯浩,李金榜. 小麦主要农艺性状的相关性及聚类分析[J]. 作物杂志, 2018, (3): 57–60 |
| [14] | 姜丽娜,岳影,李金娜,张雅雯,朱娅林,李春喜. 施氮量对小麦花后氮素分配及氮素利用的影响[J]. 作物杂志, 2018, (2): 80–86 |
| [15] | 石晓华,杨海鹰,康文钦,秦永林,樊明寿,贾立国. 不同施氮量对马铃薯-小麦轮作体系产量及土壤氮素平衡的影响[J]. 作物杂志, 2018, (2): 108–116 |
|
||