作物杂志,2021, 第3期: 51–56 doi: 10.16035/j.issn.1001-7283.2021.03.008
四川豌豆种质资源白粉病抗性及分子鉴定
项超1( ), 孙素丽2, 朱振东2, 宗绪晓2(
), 孙素丽2, 朱振东2, 宗绪晓2( ), 杨涛2, 刘荣2, 杨梅1, 鲜东锋1, 杨秀燕1
), 杨涛2, 刘荣2, 杨梅1, 鲜东锋1, 杨秀燕1
- 1四川省农业科学院作物研究所,610066,四川成都
2中国农业科学院作物科学研究所,100081,北京
Resistance and Molecular Identification to Powdery Mildew of Pea Germplasms in Sichuan
Xiang Chao1( ), Sun Suli2, Zhu Zhendong2, Zong Xuxiao2(
), Sun Suli2, Zhu Zhendong2, Zong Xuxiao2( ), Yang Tao2, Liu Rong2, Yang Mei1, Xian Dongfeng1, Yang Xiuyan1
), Yang Tao2, Liu Rong2, Yang Mei1, Xian Dongfeng1, Yang Xiuyan1
- 1Crop Research Institute, Sichuan Academy of Agricultural Sciences, Chengdu 610066, Sichuan, China
2Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
摘要:
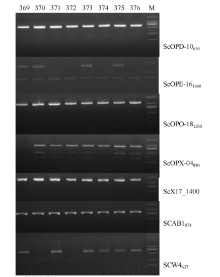
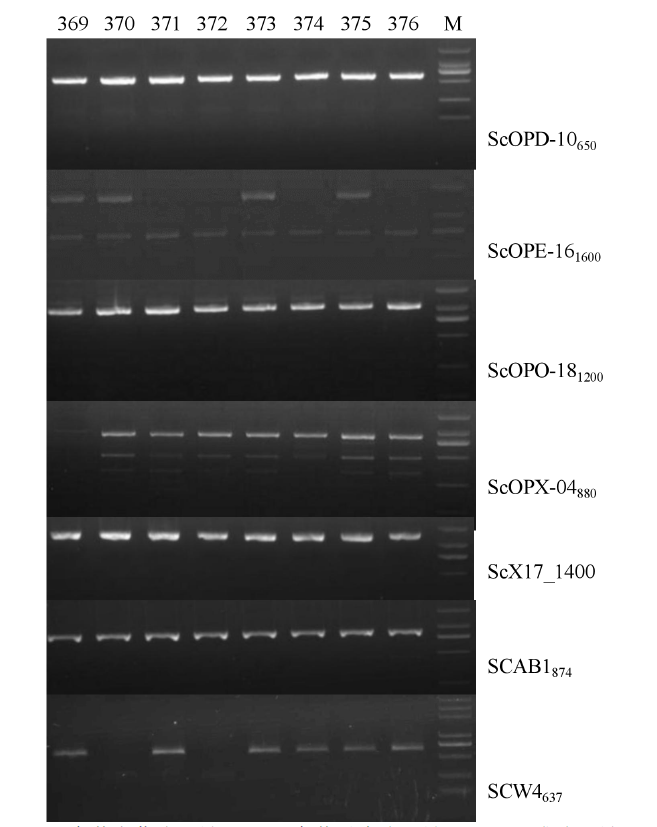
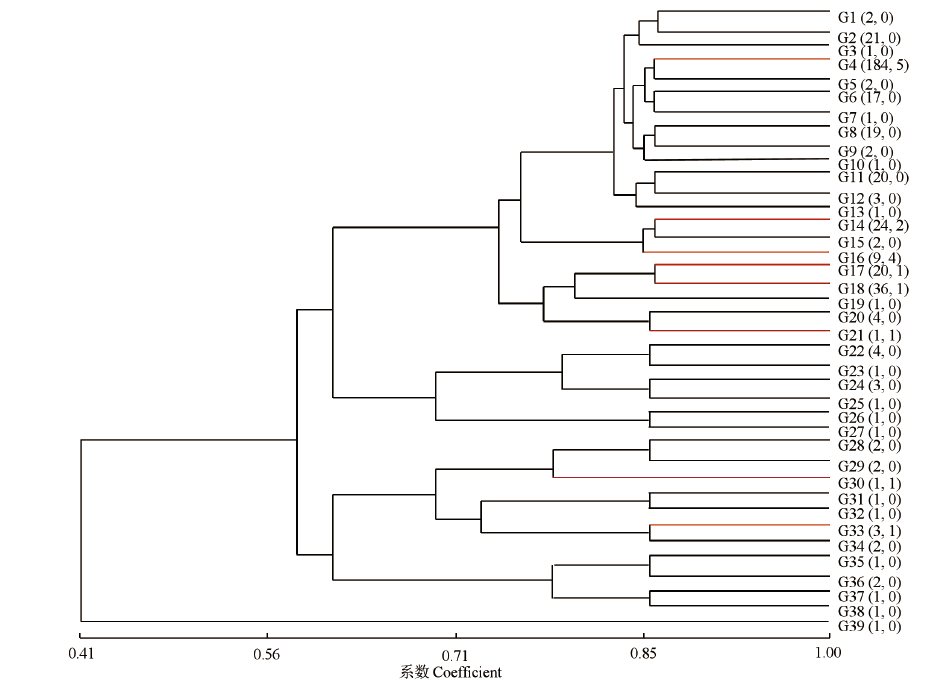
豌豆白粉病是由白粉菌(Erysiphe pisi D. C.)引起的豌豆最重要的病害之一,控制豌豆白粉病最经济有效且环保的方法是种植抗病品种。在温室条件下对400份豌豆种质资源进行白粉病抗性鉴定,同时利用7个与已知豌豆抗白粉病基因连锁的分子标记进行基因型鉴定。结果表明,在400份资源中,有8份表现免疫,3份表现抗病,5份表现抗感分离,其余384份均为感病;16份抗性资源中有10份来自于四川省中部不同纬度地区,其余6份为国外引进资源;7个分子标记将400份种质资源分为39个标记基因型,其中16份抗性资源分为7个标记基因型。上述抗性资源及其标记基因型可有效应用于豌豆白粉病抗性育种的研究中。
| [1] | Faostat. . Data Crops. (2020-12-22)[2020-12-29]. http://www.fao.org/faostat/en/#data/QC. |
| [2] |
Warkentin T, Rashid K, Xue A . Fungicidal control of powdery mildew in field pea. Canadian Journal of Plant Science, 1996,76(4):933-935.
doi: 10.4141/cjps96-156 |
| [3] |
Fondevilla S, Cubero J, Rubiales D . Confirmation that the Er3 gene,conferring resistance to Erysiphe pisi in pea,is a different gene from er1 and er2 genes. Plant Breeding, 2011,130(2):281-282.
doi: 10.1111/pbr.2011.130.issue-2 |
| [4] |
Ghafoor A, McPhee K . Marker assisted selection (MAS) for developing powdery mildew resistant pea cultivars. Euphytica, 2012,186(3):593-607.
doi: 10.1007/s10681-011-0596-6 |
| [5] |
Sun S, Fu H, Wang Z , et al. Discovery of a novel er1 allele conferring powdery mildew resistance in Chinese pea (Pisum sativum L.) landraces. PLoS ONE, 2016,11(1):e0147624.
doi: 10.1371/journal.pone.0147624 |
| [6] |
Harland S . Inheritance of immunity to mildew in Peruvian forms of Pisum sativum. Heredity, 1948,2(Pt 2):263-269.
doi: 10.1038/hdy.1948.15 |
| [7] |
Heringa R, Norel A V, Tazelaar M . Resistance to powdery mildew (Erisyphe polygoni DC) in peas (Pisum sativum L.). Euphytica, 1969,18(2):163-169.
doi: 10.1007/BF00035687 |
| [8] |
Fondevilla S, Carver T L W, Moreno M T , et al. Macroscopic and histological characterisation of genes er1 and er2 for powdery mildew resistance in pea. European Journal of Plant Pathology, 2006,115(3):309-321.
doi: 10.1007/s10658-006-9015-6 |
| [9] |
Fondevilla S, Torres A M, Moreno M T , et al. Identification of a new gene for resistance to powdery mildew in Pisum fulvum,a wild relative of pea. Breeding Science, 2007,57(2):181-184.
doi: 10.1270/jsbbs.57.181 |
| [10] | Ondřej M, Dostálová R, Odstrčilová L . Response of Pisum sativum germplasm resistant to Erysiphe pisi to inoculation with Erysiphe baeumleri,a new pathogen of peas. Plant Protection Science, 2005(41):95-103. |
| [11] |
Attanayake R N, Glawe D A, Mcphee K E , et al. Erysiphe trifolii-a newly recognized powdery mildew pathogen of pea. Plant Pathology, 2010,59(4):712-720.
doi: 10.1111/ppa.2010.59.issue-4 |
| [12] |
Fondevilla S, Chattopadhyay C, Khare N , et al. Erysiphe trifolii is able to overcome er1 and Er3,but not er2,resistance genes in pea. European Journal of Plant Pathology, 2013,136(3):557-563.
doi: 10.1007/s10658-013-0187-6 |
| [13] |
Tiwari K R, Penner G A, Warkentin T D , et al. Pathogenic variation in Erysiphe pisi,the causal organism of powdery mildew of pea. Canadian Journal of Plant Pathology, 1997,19(3):267-271.
doi: 10.1080/07060669709500522 |
| [14] |
Tiwari K R, Penner G A, Warkentin T D . Inheritance of powdery mildew resistance in pea. Canadian Journal of Plant Science, 1997,77(3):307-310.
doi: 10.4141/P96-157 |
| [15] |
Fondevilla S, Rubiales D . Powdery mildew control in pea. A review. Agronomy for Sustainable Development, 2012,32(2):401-409.
doi: 10.1007/s13593-011-0033-1 |
| [16] |
Sun S, Wang Z, Fu H , et al. Resistance to powdery mildew in the pea cultivar Xucai 1 is conferred by the gene er1. The Crop Journal, 2015,3(6):489-499.
doi: 10.1016/j.cj.2015.07.006 |
| [17] |
Timmerman G M, Frew T J, Weeden N F , et al. Linkage analysis of er-1,a recessive Pisum sativum gene for resistance to powdery mildew fungus (Erysiphe pisi D.C.). Theoretical and Applied Genetics, 1994,88(8):1050-1055.
doi: 10.1007/BF00220815 pmid: 24186261 |
| [18] |
Tiwari K, Penner G, Warkentin T . Identification of coupling and repulsion phase RAPD markers for powdery mildew resistance gene er-1 in pea. Genome, 1998,41(3):440-444.
doi: 10.1139/g98-014 |
| [19] |
Srivastava R K, Mishra S K, Singh A K , et al. Development of a coupling-phase SCAR marker linked to the powdery mildew resistance gene ‘er1’ in pea (Pisum sativum L.). Euphytica, 2012,186(3):855-866.
doi: 10.1007/s10681-012-0650-z |
| [20] |
Katoch V, Sharma S, Pathania S , et al. Molecular mapping of pea powdery mildew resistance gene er2 to pea linkage group Ⅲ. Molecular Breeding, 2010,25(2):229-237.
doi: 10.1007/s11032-009-9322-7 |
| [21] |
Fondevilla S, Rubiales D, Moreno M T , et al. Identification and validation of RAPD and SCAR markers linked to the gene Er3 conferring resistance to Erysiphe pisi DC in pea. Molecular Breeding, 2008,22(2):193-200.
doi: 10.1007/s11032-008-9166-6 |
| [22] |
Kreplak J, Madoui M-A, Cápal P , et al. A reference genome for pea provides insight into legume genome evolution. Nature Genetics, 2019,51(9):1411-1422.
doi: 10.1038/s41588-019-0480-1 |
| [23] |
Kulaeva O A, Zhernakov A I, Afonin A M , et al. Pea Marker Database (PMD) - A new online database combining known pea (Pisum sativum L.) gene-based markers. PLoS ONE, 2017,12(10):e0186713.
doi: 10.1371/journal.pone.0186713 |
| [24] | 彭化贤, 姚革, 贾瑞林 , 等. 豌豆抗白粉病资源鉴定研究. 西南农业大学学报, 1991(4):20-22. |
| [25] | 曾亮, 李敏权, 杨晓明 . 豌豆种质资源白粉病抗性鉴定. 草原与草坪, 2012,32(4):35-38. |
| [26] | 陆建英, 杨晓明, 王昶 , 等. 抗白粉病豌豆种质资源田间筛选. 植物保护, 2015(3):154-158. |
| [27] | 王仲怡, 包世英, 段灿星 , 等. 豌豆抗白粉病资源筛选及分子鉴定. 作物学报, 2013,39(6):1030-1038. |
| [28] | 付海宁, 孙素丽, 朱振东 , 等. 加拿大豌豆品种(系)抗白粉病表型和基因型鉴定. 植物遗传资源学报, 2014,15(5):1028-1033. |
| [29] |
Pavan S, Schiavulli A, Appiano M , et al. Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theoretical and Applied Genetics, 2011,123(8):1425-1431.
doi: 10.1007/s00122-011-1677-6 |
| [30] |
Humphry M, Reinstaedler A, Ivanov S , et al. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Molecular Plant Pathology, 2011,12(9):866-878.
doi: 10.1111/j.1364-3703.2011.00718.x pmid: 21726385 |
| [31] |
Santo T, Rashkova M, Alabaça C , et al. The ENU-induced powdery mildew resistant mutant pea (Pisum sativum L.) lines S (er1mut1) and F (er1mut2) harbour early stop codons in the PsMLO1 gene. Molecular breeding, 2013,32(3):723-727.
doi: 10.1007/s11032-013-9889-x |
| [32] |
Sun S, Deng D, Wang Z , et al. A novel er1 allele and the development and validation of its functional marker for breeding pea (Pisum sativum L.) resistance to powdery mildew. Theoretical and Applied Genetics, 2016,129(5):909-919.
doi: 10.1007/s00122-016-2671-9 |
| [33] | 王仲怡, 付海宁, 孙素丽 , 等. 豌豆品系X9002抗白粉病基因鉴定. 作物学报, 2015,41(4):515-523. |
| [34] |
Liu S M, O'Brien L, Moore S G , et al. A single recessive gene confers effective resistance to powdery mildew of field pea grown in northern New South Wales. Australian Journal of Experimental Agriculture, 2003,43(4):373-378.
doi: 10.1071/EA01142 |
| [35] |
Ek M, Eklund M, Von P R , et al. Microsatellite markers for powdery mildew resistance in pea (Pisum sativum L.). Hereditas, 2005,142(2005):86-91.
pmid: 16970617 |
| [1] | 李琼, 常世豪, 武婷婷, 耿臻, 杨青春, 舒文涛, 李金花, 张东辉, 张保亮. 120份大豆种质资源遗传多样性和亲缘关系分析[J]. 作物杂志, 2021, (4): 51–58 |
| [2] | 贾瑞玲, 赵小琴, 南铭, 陈富, 刘彦明, 魏立平, 刘军秀, 马宁. 64份苦荞种质资源农艺性状遗传多样性分析与综合评价[J]. 作物杂志, 2021, (3): 19–27 |
| [3] | 李梦钰, 高闯, 李巧云, 徐凯歌, 王丝雨, 牛吉山. 苗期及灌浆期抗Bipolaris sorokiniana叶枯病小麦品种(系)鉴定及相关性分析[J]. 作物杂志, 2021, (3): 40–45 |
| [4] | 潘晓雪, 胡明瑜, 王忠伟, 吴红, 雷开荣. 不同水稻种质资源重要农艺性状与发芽期耐寒性鉴定研究[J]. 作物杂志, 2021, (1): 47–53 |
| [5] | 曾艳华, 谢和霞, 江禹奉, 周锦国, 谢小东, 周海宇, 谭贤杰, 覃兰秋, 程伟东. 基于SNP标记的爆裂玉米农家品种遗传多样性[J]. 作物杂志, 2020, (5): 65–70 |
| [6] | 宫彦龙, 雷月, 闫志强, 刘雪薇, 张大双, 吴健强, 朱速松. 不同生态区粳稻资源表型遗传多样性综合评价[J]. 作物杂志, 2020, (5): 71–79 |
| [7] | 陈卫国, 张政, 史雨刚, 曹亚萍, 王曙光, 李宏, 孙黛珍. 211份小麦种质资源抗旱性的评价[J]. 作物杂志, 2020, (4): 53–63 |
| [8] | 齐冰洁, 王敏, 张智勇, 贺鑫, 刘景辉. 燕麦种质资源矿质元素的多样性分析[J]. 作物杂志, 2020, (4): 72–78 |
| [9] | 公丹, 王素华, 程须珍, 王丽侠. 普通豇豆应用核心种质的SSR指纹图谱构建及多样性分析[J]. 作物杂志, 2020, (4): 79–83 |
| [10] | 刘栋, 马琴, 李爱荣, 郭娜, 马建富, 乔海明, 苗红梅. 亚麻种质资源种子形态性状与含油量的分析与评价[J]. 作物杂志, 2020, (3): 34–41 |
| [11] | 刘歆,朱容,杨梅,刘章勇. 再生稻高产种质资源筛选及高产群体分析[J]. 作物杂志, 2020, (2): 28–33 |
| [12] | 孙瑞东,臧振原,慈佳宾,杨巍,任雪娇,姜良宇,杨伟光. 玉米自交系对大斑病菌的抗性鉴定及抗性来源分析[J]. 作物杂志, 2020, (2): 65–70 |
| [13] | 李松,张世成,董云武,施德林,史云东. 基于SSR标记的云南腾冲水稻的遗传多样性分析[J]. 作物杂志, 2019, (5): 15–21 |
| [14] | 吕伟,韩俊梅,任果香,文飞,王若鹏,刘文萍. 山西芝麻种质资源遗传多样性分析[J]. 作物杂志, 2019, (5): 57–63 |
| [15] | 李晶,南铭. 俄罗斯和乌克兰引进冬小麦在我国西北地区的农艺性状表现和遗传多样性分析[J]. 作物杂志, 2019, (5): 9–14 |
|
||