作物杂志,2021, 第5期: 64–71 doi: 10.16035/j.issn.1001-7283.2021.05.010
基于SNP分子标记的泰山/泰科麦系列小麦遗传解析
亓晓蕾1( ), 李兴锋2(
), 李兴锋2( ), 吕广德1, 王瑞霞1, 王君3, 孙宪印1, 孙盈盈1, 陈永军1, 钱兆国1(
), 吕广德1, 王瑞霞1, 王君3, 孙宪印1, 孙盈盈1, 陈永军1, 钱兆国1( ), 吴科1
), 吴科1
- 1泰安市农业科学院,271000,山东泰安
2国家小麦改良中心泰安分中心,271018,山东泰安
3泰安市农业农村局,山东泰安,271000
Genetic Analysis of Taishan/Taikemai Serial Wheat Based on SNP Molecular Markers
Qi Xiaolei1( ), Li Xingfeng2(
), Li Xingfeng2( ), Lü Guangde1, Wang Ruixia1, Wang Jun3, Sun Xianyin1, Sun Yingying1, Chen Yongjun1, Qian Zhaoguo1(
), Lü Guangde1, Wang Ruixia1, Wang Jun3, Sun Xianyin1, Sun Yingying1, Chen Yongjun1, Qian Zhaoguo1( ), Wu Ke1
), Wu Ke1
- 1Tai’an Academy of Agricultural Sciences, Tai’an 271000, Shandong, China
2Tai’an Subcenter of National Wheat Improvement Center, Tai’an 271018, Shandong, China
3Tai’an Agriculture and Rural Bureau, Tai’an 271000, Shandong, China
摘要:
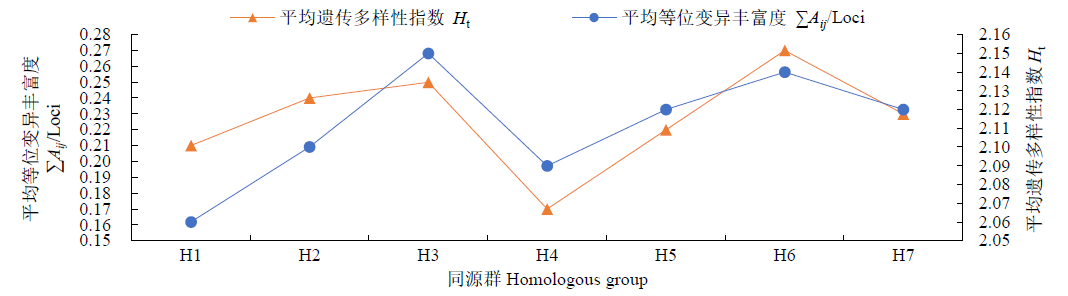
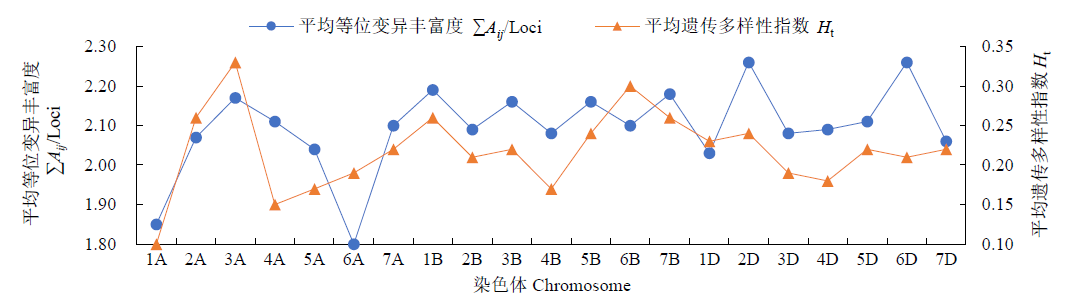
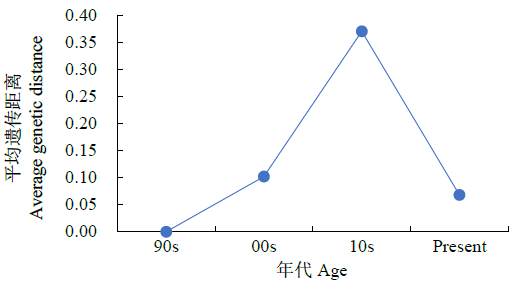
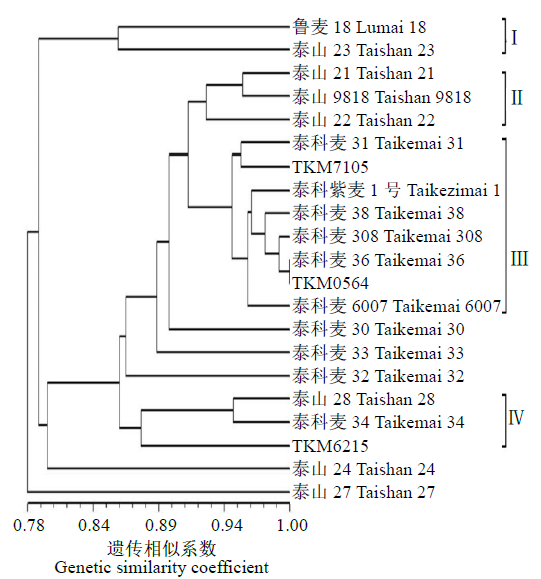
对不同年份育成的21个小麦品种(系)进行全基因组扫描,通过分析遗传距离和染色体区段/位点,明确其亲缘关系远近和遗传差异。分析可知,获得的2029个SNP基因位点在B基因组拥有较高的遗传多样性,其次是A和D基因组;在7个同源群中,第3和第6同源群呈现出较高的遗传多样性,而第1和第4同源群的遗传多样性较低;21条染色体中,3A、1B、6B染色体的遗传多样性较高,而1A、6A的遗传多样性偏低。对21份供试材料依据审定(育成)年份分析其群体的平均遗传距离,不同年份品种间的平均遗传距离先增大后减小,遗传多样性逐渐降低;21份供试材料间的遗传相似系数在0.69~0.99之间,大致可聚为4个类群,同一年份的品种一般聚在一起,与其系谱关系吻合。构建并分析供试材料的基因型图谱发现,00s、10s和现在育成的小麦品种(系)共有SNP和共有染色体区段分别主要在A、D和B基因组,对应已发表性状同不同年份育种目标吻合。同时发现21份供试材料均含有25个共同SNP位点,分布在1A、5A、6A、7A、2B、3B、6B、1D、2D、3D和7D染色体上,且每条染色体上分布的SNP位点数目均不相同,通过对应已发表性状进一步证实在品种(系)组配与选育过程中注重产量、株高、分蘖数、抽穗期、灌浆速率和抗病等性状的选择。以上研究结果可为今后小麦新品种组配和选育提供参考依据。
| [1] | 刘志勇, 王道文, 张爱民, 等. 小麦育种行业创新现状与发展趋势. 植物遗传资源学报, 2018, 19(3):430-434. |
| [2] | 吕晓欢, 冯晶, 蔺瑞明, 等. 2009-2010年河南、河北、山东三省小麦区试品种(系)的亲缘系数分析. 麦类作物学报, 2013, 33(3):455-460. |
| [3] | 蒲艳艳, 宫永超, 李娜娜, 等. 中国小麦作物遗传多样性研究进展. 中国农学通报, 2016, 32(30):7-13. |
| [4] |
Sun Q X, Ni Z F, Liu Z Y, et al. Genetic relationships and diversity among Tibetan wheat,common wheat and European spelt wheat revealed by RAPD markers. Euphytica, 1998, 99:205-211.
doi: 10.1023/A:1018316129246 |
| [5] | Talebi R, Fayyaz F. Quantitative evaluation of genetic diversity in Iranian modern cultivars of wheat (Triticum aestivum L.) using morphological and amplified fragment length polymorphism (AFLP) markers. Biharean Biologist, 2012, 6(1):14-18. |
| [6] | 王江春, 胡延吉, 余松烈, 等. 建国以来山东省小麦品种及其亲本的亲缘系数分析. 中国农业科学, 2006, 39(4):664-672. |
| [7] |
Wang H Y, Wang X E, Chen P D, et al. Assessment of genetic diversity of Yunnan,Tibetan,and Xinjiang wheat using SSR markers. Journal of Genetic and Genomics, 2007, 34:623-633.
doi: 10.1016/S1673-8527(07)60071-X |
| [8] | 傅晓艺, 刘桂茹, 杨学举. 中国部分优质小麦品种(系) 遗传多样性的SSR 分析. 麦类作物学报, 2007, 27(5):776-780. |
| [9] | 蒲艳艳, 程凯, 李斯深. 山东省近期育成小麦品种遗传多样性的SSR 分析. 分子植物育种, 2011, 9(4):443-449. |
| [10] |
Colasuonno P, Gadaleta A, Giancaspro A, et al. Development of a high- density SNP- based linkage map and detection of yellow pigment content QTLs in durum wheat. Molecular Breeding, 2014, 34(4):1563-1578.
doi: 10.1007/s11032-014-0183-3 |
| [11] |
Allen A M, Winfield M O, Burridge A J, et al. Characterization of a wheat breeder’s array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnology Journal, 2017, 15(3):390-401.
doi: 10.1111/pbi.2017.15.issue-3 |
| [12] |
Avni R, Nave M, Eilam T, et al. Ultra-dense genetic map of durum wheat × wild emmer wheat developed using the 90K iSelect SNP genotyping assay. Molecular Breeding, 2014, 34(4):1549-1562.
doi: 10.1007/s11032-014-0176-2 |
| [13] | Lucas S J, Salantur A, Yazar S, et al. High-throughput SNP genotyping of modern and wild emmer wheat for yield and root morphology using a combined association and linkage analysis. Functional and Integrative Genomics, 2017, 6(17):667-685. |
| [14] | 李玉刚, 任民, 孙绿, 等. 利用SSR 和SNP 标记分析鲁麦14 对青农2 号的遗传贡献. 作物学报, 2018, 44(2):159-168. |
| [15] |
Xu X T, Zhu Z W, Jia A L, et al. Mapping of QTL for partial resistance to powdery mildew in two Chinese common wheat cultivars. Euphytica, 2020, 216:3.
doi: 10.1007/s10681-019-2537-8 |
| [16] |
Tan M K, EI- Bouhssini M, Emebiri L, et al. A SNP marker for the selection of HfrDrd,a Hessian fly- response gene in wheat. Molecular Breeding, 2015, 35:216-225.
doi: 10.1007/s11032-015-0410-6 |
| [17] |
Lin M, Cai S B, Wang S, et al. Genotyping-by-sequencing (GBS)identified SNP tightly linked to QTL for pre-harvest sprouting resistance. Theoretical and Applied Genetics, 2015, 128(7):1385-1395.
doi: 10.1007/s00122-015-2513-1 |
| [18] |
Gao L L, Kielsmeier-Cook J, Bajgain P, et al. Development of genotyping by sequencing (GBS)- and array-derived SNP markers for stem rust resistance gene Sr42. Molecular Breeding, 2015, 35:207.
doi: 10.1007/s11032-015-0404-4 |
| [19] |
Liu W Z, Maccaferri M, Bulli P, et al. Genome-wide association mapping for seedling and field resistance to Puccinia striiformis f.sp. tritici in elite durum wheat. Theoretical and Applied Genetics, 2017, 130(4):649-667.
doi: 10.1007/s00122-016-2841-9 |
| [20] |
Jighly A, Oyiga B C, Makdis F, et al. Genome-wide DArT and SNP scan for QTL associated with resistance to stripe rust (Puccinia striiformis f. sp. tritici) in elite ICARDA wheat (Triticum aestivum L.) germplasm. Theoretical and Applied Genetics, 2015, 128(7):1277-1295.
doi: 10.1007/s00122-015-2504-2 |
| [21] |
Chen G F, Zhang H, Deng Z Y, et al. Genome-wide association study for kernel weight-related traits using SNPs in a Chinese winter wheat population. Euphytica, 2016, 212(2):173-185.
doi: 10.1007/s10681-016-1750-y |
| [22] | 翟俊鹏, 李海霞, 毕惠惠, 等. 普通小麦主要农艺性状的全基因组关联分析. 作物学报, 2019, 45(10):1488-1502. |
| [23] | 曹廷杰, 谢菁忠, 吴秋红, 等. 河南省近年审定小麦品种基于系谱和SNP 标记的遗传多样性分析. 作物学报, 2015, 41(2):197-206. |
| [24] |
Mangini G, Margiotta B, Marcotuli I, et al. Genetic diversity and phenetic analysis in wheat (Triticum turgidum subsp. durum and Triticum aestivum subsp. aestivum) landraces based on SNP markers. Genetic Resources and Crop Evolution, 2017, 64(6):1269-1280.
doi: 10.1007/s10722-016-0435-7 |
| [25] | 刘易科, 朱展望, 陈泠, 等. 基于SNP标记揭示我国小麦品种(系)的遗传多样性. 作物学报, 2020, 46(2):307-314. |
| [26] |
Chao S M, Dubcovsky J, Dvorak J, et al. Population- and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.). BMC Genomics, 2010, 11:727.
doi: 10.1186/1471-2164-11-727 |
| [27] | 郝晨阳, 王兰芬, 张学勇, 等. 我国育成小麦品种的遗传多样性演变. 中国科学 C 辑, 2005, 35(5):408-415. |
| [28] | Rohlf F J. NTSYS-pc:Numerical taxonomy and multivariate analysis system. Version 2.2. New York:Exeter Software, 2009. |
| [29] |
Zhang Q F, Allard R W. Sampling variance of the genetic diversity index. Journal of Heredity, 1986, 77:54-55.
doi: 10.1093/oxfordjournals.jhered.a110169 |
| [30] |
Ren J, Chen L, Sun D K, et al. SNP-revealed genetic diversity in wild emmer wheat correlates with ecological factors. BMC Evolutionary Biology, 2013, 13:169.
doi: 10.1186/1471-2148-13-169 |
| [31] |
Würschum T, Langer S M, Longin C F H, et al. Population structure,genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theoretical and Applied Genetics, 2013, 126:1477-1486.
doi: 10.1007/s00122-013-2065-1 pmid: 23429904 |
| [32] | 盖红梅, 李玉刚, 王瑞英, 等. 鲁麦14 对山东新选育小麦品种的遗传贡献. 作物学报, 2012, 38(6):954-961. |
| [33] | 程斌, 张淑英, 张明霞, 等. 山东省近年育成小麦品种(系)的遗传多样性分析. 山东农业科学, 2016, 48(9) :17-22. |
| [34] |
Su J Y, Zheng Q, Li H W, et al. Detection of QTLs for phosphorus use efficiency in relation to agronomic performance of wheat grown under phosphorus sufficient and limited conditions. Plant Science, 2009, 176:824-836.
doi: 10.1016/j.plantsci.2009.03.006 |
| [35] | 贾继增. 小麦分子标记研究进展. 生物技术通报, 1997(2):1-5. |
| [36] |
Jia H Y, Wan H S, Yang S H, et al. Genetic dissection of yield-related traits in a recombinant inbred line population created using a key breeding parent in China’s wheat breeding. Theoretical and Applied Genetics, 2013, 126(8):2123-2139.
doi: 10.1007/s00122-013-2123-8 |
| [37] |
Wang R X, Hai L, Zhang X Y, et al. QTL mapping for grain filling rate and yield-related traits in RILs of the Chinese winter wheat population Heshangmai × Yu8679. Theoretical and Applied Genetics, 2009, 118(2):313-325.
doi: 10.1007/s00122-008-0901-5 pmid: 18853131 |
| [38] |
Narasimhamoorthy B, Gill B S, Fritz A K, et al. Advanced backcross QTL analysis of a hard winter wheat × synthetic wheat population. Theoretical and Applied Genetics, 2006, 112(5):787-796.
pmid: 16463062 |
| [39] |
Kumar N, Kulwal P L, Balyan H S, et al. QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Molecular Breeding, 2007, 19:163-177.
doi: 10.1007/s11032-006-9056-8 |
| [40] |
Zhang W, Gianibelli M C, Rampling L R, et al. Characterization and marker development for low molecular weight glutenin genes from Glu-A3 alleles of bread wheat (Triticum aestivum L.). Theoretical and Applied Genetics, 2004, 108(7):1409-1419.
pmid: 14727031 |
| [41] |
Gupta P K, Balyan H S, Sharma S, et al. Genetics of yield,abiotic stress tolerance and biofortification in wheat (Triticum aestivum L.). Theoretical and Applied Genetics, 2020, 133(3):1569-1602.
doi: 10.1007/s00122-020-03583-3 |
| [42] |
Chen W G, Sun D Z, Yan X, et al. QTL analysis of wheat kernel traits,and genetic effects of qKW-6A on kernel width. Euphytica, 2019, 215:11.
doi: 10.1007/s10681-018-2333-x |
| [43] |
Hai L, Guo H J, Wagner C, et al. Genomic regions for yield and yield parameters in Chinese winter wheat (Triticum aestivum L.) genotypes tested under varying environments correspond to QTL in widely different wheat materials. Plant Science, 2008, 175:226-232.
doi: 10.1016/j.plantsci.2008.03.006 |
| [44] | Zhai H J, Feng Z Y, Li J, et al. QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Frontiers in Plant Science, 2016, 7:1617. |
| [45] |
Cuthbert J L, Somers D J, Brûlé-Babel A L, et al. Molecular mapping of quantitative trait loci for yield and yield components in spring wheat (Triticum aestivum L.). Theoretical and Applied Genetics, 2008, 117(4):595-608.
doi: 10.1007/s00122-008-0804-5 pmid: 18516583 |
| [46] |
Reif J C, Maurer H P, Korzun V, et al. Mapping QTLs with main and epistatic effects underlying grain yield and heading time in soft winter wheat. Theoretical and Applied Genetics, 2011, 123(2):283-292.
doi: 10.1007/s00122-011-1583-y |
| [47] |
Chen Z Y, Cheng X J, Chai L L, et al. Pleiotropic QTL influencing spikelet number and heading date in common wheat (Triticum aestivum L.). Theoretical and Applied Genetics, 2020, 133(1):1825-1838.
doi: 10.1007/s00122-020-03556-6 |
| [48] |
Huang X Q, Cloutier S, Lycar L, et al. Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.). Theoretical and Applied Genetics, 2006, 113(4):753-766.
pmid: 16838135 |
| [49] |
Zhang P P, Lan C X, Asad M A, et al. QTL mapping of adult-plant resistance to leaf rust in the Chinese landraces Pingyuan 50/Mingxian 169 using the wheat 55K SNP array. Molecular Breeding, 2019, 39:98.
doi: 10.1007/s11032-019-1004-5 |
| [50] |
Carter A H, Garland-Campbell K, Kidwell K K. Genetic mapping of quantitative trait loci associated with Important agronomic traits in the spring wheat (Triticum aestivum L.) cross ‘Louise’ × ‘Penawawa’. Crop Science, 2011, 51:84-95.
doi: 10.2135/cropsci2010.03.0185 |
| [51] |
Nakamura T, Yamamori M, Hirano H, et al. Identification of three Wx proteins in wheat (Triticum aestivum L.). Biochemical Genetics, 1993, 31:75-86.
pmid: 8471025 |
| [52] |
He X Y, He Z H, Zhang L P, et al. Allelic variation of polyphenol oxidase (PPO) genes located on chromosomes 2A and 2D and development of functional markers for the PPO genes in common wheat. Theoretical and Applied Genetics, 2007, 115(1):47-58.
pmid: 17426955 |
| [53] | 赵春华, 樊小莉, 王维莲, 等. 小麦候选骨干亲本科农9204 遗传构成及其传递率. 作物学报, 2015, 41(4):574-584. |
| [54] |
Zhang D L, Hao C Y, Wang L F, et al. Identifying loci influencing grain number by microsatellite screening in bread wheat (Triticum aestivum L.). Planta, 2012, 236:1507-1517.
doi: 10.1007/s00425-012-1708-9 |
| [55] |
Huang X Q, Kempf H, Ganal M W, et al. Advanced backcross QTL analysis in progenies derived from a cross between a German elite winter wheat variety and a synthetic wheat (Triticum aestivum L.). Theoretical and Applied Genetics, 2004, 109(5):933-943.
pmid: 15243706 |
| [56] |
Ollier M, Talle V, Brisset A L, et al. QTL mapping and successful introgression of the spring wheat‑derived QTL Fhb1 for Fusarium head blight resistance in three European triticale populations. Theoretical and Applied Genetics, 2020, 133(1):457-477.
doi: 10.1007/s00122-019-03476-0 |
| [57] |
Sun X C, Marza F, Ma H X, et al. Mapping quantitative trait loci for quality factors in an inter-class cross of US and Chinese wheat. Theoretical and Applied Genetics, 2010, 120(5):1041-1051.
doi: 10.1007/s00122-009-1232-x |
| [58] |
Sun X Y, Wu K, Zhao Y, et al. QTL analysis of kernel shape and weight using recombinant inbred lines in wheat. Euphytica, 2009, 165:615-624.
doi: 10.1007/s10681-008-9794-2 |
| [1] | 吴鑫雨, 刘振洋, 李海叶, 郑毅, 汤利, 肖靖秀. 施氮和间作对蚕豆根瘤形成及氮素吸收累积的影响[J]. 作物杂志, 2021, (5): 120–127 |
| [2] | 段振盈, 徐新玉, 李星, 李在峰, 马骏, 姚占军. 12个主产区历史小麦品种抗叶锈病基因分析[J]. 作物杂志, 2021, (5): 20–27 |
| [3] | 王云, 乔玲, 闫素仙, 武棒棒, 郑兴卫, 赵佳佳. 山西省不同年代旱地小麦产量构成因素及抗旱性特征分析[J]. 作物杂志, 2021, (5): 43–49 |
| [4] | 刘歆, 杨芳, 邓军波, 汪嫒嫒, 何念, 陈艳. 江汉平原大豆品系表型分析及综合评价[J]. 作物杂志, 2021, (5): 57–63 |
| [5] | 张全芳, 姜明松, 陈峰, 朱文银, 周学标, 杨连群, 徐建第. 山东省水稻品种(系)的遗传多样性分析[J]. 作物杂志, 2021, (4): 26–31 |
| [6] | 杜晓宇, 李楠楠, 邹少奎, 王丽娜, 吕永军, 张倩, 李顺成, 杨光宇, 韩玉林. 黄淮南片新育成小麦品种(系)主要性状的综合性分析[J]. 作物杂志, 2021, (4): 38–45 |
| [7] | 李梦钰, 高闯, 李巧云, 徐凯歌, 王丝雨, 牛吉山. 苗期及灌浆期抗Bipolaris sorokiniana叶枯病小麦品种(系)鉴定及相关性分析[J]. 作物杂志, 2021, (3): 40–45 |
| [8] | 周正萍, 田宝庚, 陈婉华, 王子阳, 袁伟, 刘世平. 不同耕作方式与秸秆还田对土壤养分及小麦产量和品质的影响[J]. 作物杂志, 2021, (3): 78–83 |
| [9] | 赵庆玲, 林文, 任爱霞, 张蓉蓉, 李蕾, 孙敏, 高志强. 春季追肥对冬小麦群体构建和籽粒灌浆进程的影响[J]. 作物杂志, 2021, (3): 99–105 |
| [10] | 贾子苗, 邱玉亮, 林志珊, 王轲, 叶兴国. 利用近缘种属优良基因改良小麦研究进展[J]. 作物杂志, 2021, (2): 1–14 |
| [11] | 刘阿康, 王德梅, 王艳杰, 杨玉双, 马瑞琦, 高甜甜, 王玉娇, 阚茗溪, 赵广才, 常旭虹. 苗期调控对晚播小麦产量及氮素利用的影响[J]. 作物杂志, 2021, (2): 116–123 |
| [12] | 王玉娇, 曹祺, 常旭虹, 王德梅, 王艳杰, 杨玉双, 赵广才, 石书兵. 不同土壤条件下化学调控对小麦产量和品质的影响[J]. 作物杂志, 2021, (2): 96–100 |
| [13] | 刘佳敏, 汪洋, 褚旭, 齐欣, 王慢慢, 赵亚南, 叶优良, 黄玉芳. 种植密度和施氮量对小麦-玉米轮作体系下周年产量及氮肥利用率的影响[J]. 作物杂志, 2021, (1): 143–149 |
| [14] | 安娟华, 董鑫, 王克俭, 何振学. 基于GWO优化SVM的小麦籽粒优劣分级研究[J]. 作物杂志, 2021, (1): 200–206 |
| [15] | 王黎明, 孔维玮, 高华利, 董普辉, 闫雪芳, 王春平, 王洪刚, 李兴锋. 小麦4B染色体上LOX基因的等位变异及其区域分布[J]. 作物杂志, 2021, (1): 32–37 |
|
||