Crops ›› 2020, Vol. 36 ›› Issue (1): 55-60.doi: 10.16035/j.issn.1001-7283.2020.01.010
Previous Articles Next Articles
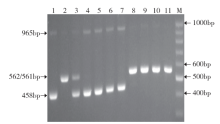
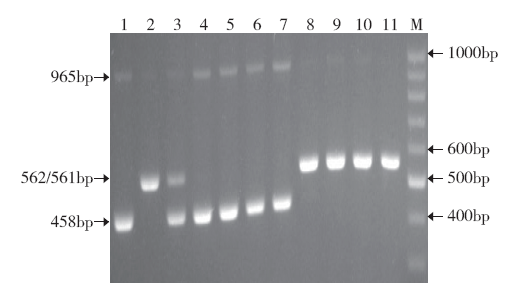
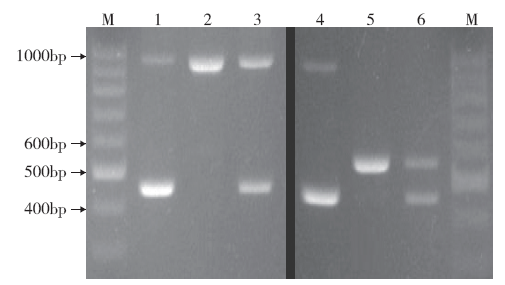
Design and Verification of New Functional Marker of Chilling-Tolerance COLD1 Gene in Rice Seedling Stage
Tian Mengxiang1,Gong Yanlong2,Zhang Shilong1( ),He Youxun1,Lei Yue2,Yu Benxun1,Yu Li1,Li Jiali2,Zhang Dashuang2,Ye Yongyin1
),He Youxun1,Lei Yue2,Yu Benxun1,Yu Li1,Li Jiali2,Zhang Dashuang2,Ye Yongyin1
- 1Bijie Institute of Agricultural Sciences, Bijie 551714, Guizhou, China
2Guizhou Rice Research Institute, Guiyang 550006, Guizhou, China
| [1] | 田孟祥, 张时龙, 余本勋 , 等. 基于四引物扩增受阻突变体系PCR快速鉴定水稻S5基因的籼粳属性. 作物杂志,2015(6):48-53. |
| [2] | Liu F X, Sun C Q, Tan L B , et al. Identification and mapping of quantitative trait loci controlling cold-tolerance of Chinese common wild rice (O. rufipogon Griff.) at booting to flowering stages. Chinese Science Bulletin, 2003,48(19):2068-2071. |
| [3] | Andaya V C, Mackill D J . QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica×indica cross. Theoretical and Applied Genetics, 2003,106(6):1084-1090. |
| [4] | Xu L M, Zhou L, Zeng Y W , et al. Identification and mapping of quantitative trait loci for cold tolerance at the booting stage in a japonica rice near-isogenic line. Plant Science, 2008,174(3):340-347. |
| [5] | Kuroki M, Saito K, Matsuba S , et al. Quantitative trait locus analysis for cold tolerance at the booting stage in a rice cultivar,hatsushizuku. Japan Agricultural Research Quarterly, 2009,43(2):115-121. |
| [6] | Lei J G, Zhu S, Shao C H , et al. Mapping quantitative trait loci for cold tolerance at the booting stage in rice by using chromosome segment substitution lines. Crop and Pasture Science, 2018,69(3):278-283. |
| [7] | Shirasawa S, Endo T, Nakagomi K , et al. Delimitation of a QTL region controlling cold tolerance at booting stage of a cultivar,'Lijiangxintuanheigu',in rice,Oryza sativa L. Theoretical and Applied Genetics, 2012,124(5):937-946. |
| [8] | Biswas P S, Khatun H, Das N , et al. Mapping and validation of QTLs for cold tolerance at seedling stage in rice from an indica cultivar Habiganj Boro VI (Hbj. BVI). 3 Biotech, 2017,7(6):359. |
| [9] | 王棋, 范淑秀, 郭江华 , 等. 利用籼粳交RIL群体对水稻发芽期和苗期耐冷性的QTL分析. 华北农学报, 2019,34(1):83-88. |
| [10] | 朱金燕, 杨梅, 嵇朝球 , 等. 利用染色体单片段代换系定位水稻芽期耐冷QTL. 植物学报, 2015,50(3):338-345. |
| [11] | 刘次桃, 王威, 毛毕刚 , 等. 水稻耐低温逆境研究:分子生理机制及育种展望. 遗传, 2018,40(3):171-185. |
| [12] | Saito K, Hayano-Saito Y, Kuroki M , et al. Map-based cloning of the rice cold tolerance gene Ctb1. Plant Science, 2010,179(1/2):97-102. |
| [13] | Kim S I, Andaya V C, Tai T H . Cold sensitivity in rice (Oryza sativa L) is strongly correlated with a naturally occurring I99V mutation in the multifunctional glutathione transferase isozyme GSTZ2. Biochemical Journal, 2011,435(2):373-380. |
| [14] | Fujino K, Sekiguchi H, Matsuda Y , et al. Molecular identification of a major quantitative trait locus,qLTG3-1,controlling low-temperature germinability in rice. Proceedings of the National Academy of Sciences of the United States of America, 2008,105(34):12623-12628. |
| [15] | Lu G W, Wu F Q, Wu W X , et al. Rice LTG1 is involved in adaptive growth and fitness under low ambient temperature. The Plant Journal, 2014,78(3):468-480. |
| [16] | Ma Y, Dai X Y, Xu Y Y , et al. COLD1 confers chilling tolerance in rice. Cell, 2015,160(6):1209-1221. |
| [17] | Zhao J, Zhang S, Dong J , et al. A novel functional gene associated with cold tolerance at the seedling stage in rice. Plant Biotechnology Journal, 2017,15(9):1141-1148. |
| [18] | Zhang Z Y, Li J J, Pan Y H , et al. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nature Communications, 2017,8:14788. |
| [19] | Liu C T, Ou S J, Mao B G , et al. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nature Communications, 2018,9(1):3302. |
| [20] | Mao D H, Xin Y Y, Tan Y J , et al. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proceedings of the National Academy of Sciences of the United States of America, 2019,116(9):3494-3501. |
| [21] | 杨佳, 曹黎明, 周继华 , 等. 水稻耐低温基因COLD1功能标记的开发及应用. 分子植物育种, 2019,17(18):6028-6032. |
| [22] | 田孟祥, 余本勋, 张时龙 , 等. 一种水稻高氮利用率NRT1.1B基因功能标记的开发与应用. 分子植物育种, 2016,14(2):410-416. |