Crops ›› 2021, Vol. 37 ›› Issue (4): 51-58.doi: 10.16035/j.issn.1001-7283.2021.04.008
Previous Articles Next Articles
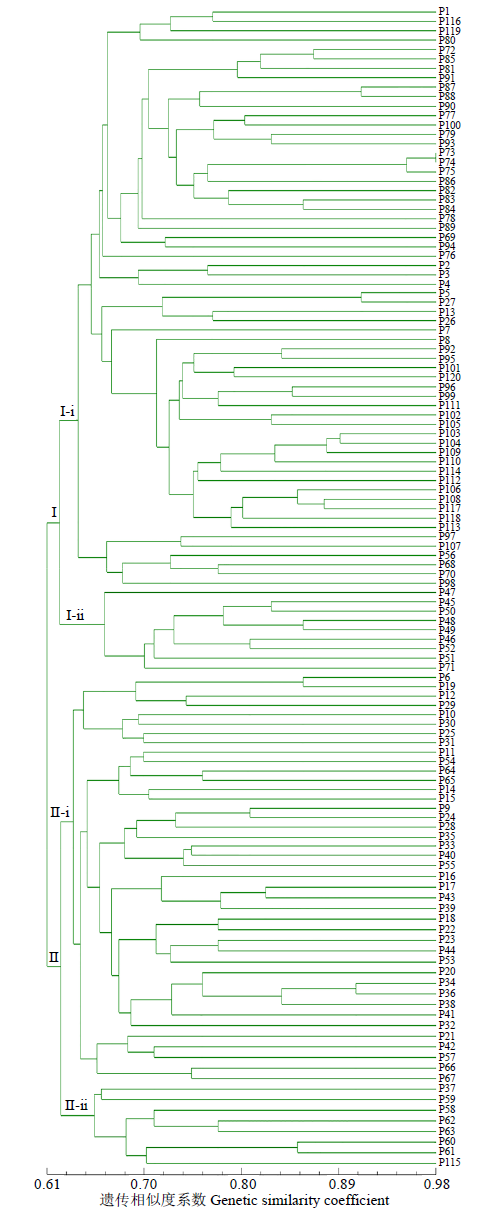
Analysis of Genetic Diversity and Genetic Relationship for 120 Soybean Germplasms
Li Qiong1( ), Chang Shihao1, Wu Tingting2, Geng Zhen1(
), Chang Shihao1, Wu Tingting2, Geng Zhen1( ), Yang Qingchun1, Shu Wentao1, Li Jinhua1, Zhang Donghui1, Zhang Baoliang1
), Yang Qingchun1, Shu Wentao1, Li Jinhua1, Zhang Donghui1, Zhang Baoliang1
- 1Zhoukou Academy of Agricultural Sciences, Zhoukou 466001, Henan, China
2Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
| [1] | 赵团结, 盖钧镒. 栽培大豆起源与演化研究进展. 中国农业科学, 2004,37(7):954-962. |
| [2] | Hymowitz T, Newell C A. Taxonomy of the genus Glycine,domestication and uses of soybeans. Economic Botany, 1981,35(3):272-288. |
| [3] | 夏正俊. 大豆基因组解析与重要农艺性状基因克隆研究进展. 植物学报, 2017,52(2):148-158. |
| [4] | 郭文韬. 略论中国栽培大豆的起源. 南京农业大学学报(社会科学版), 2004,4(1):60-69. |
| [5] | 傅旭军. 大豆种质资源遗传多样性分析及其利用研究. 杭州:浙江大学, 2009. |
| [6] | 蒲艳艳, 宫永超, 李娜娜. 中国大豆种质资源遗传多样性研究进展. 大豆科学, 2018,37(2):315-321. |
| [7] | 钟文娟, 袁灿, 周永航, 等. 基于SSR标记的四川大豆与引进大豆资源遗传多样性和群体结构分析. 大豆科学, 2017,36(5):657-668. |
| [8] | Wang L X, Guan R X, Liu Z X, et al. Genetic diversity of Chinese cultivated soybean revealed by SSR markers. Crop Science, 2006,46(3):1032-1038. |
| [9] | Wang L X, Guan R X, Liu Z X, et al. Genetic diversity of Chinese spring soybean germplasms revealed by SSR markers. Plant Breeding, 2008,127(1):56-61. |
| [10] | Iquira E, Gagnon E, Belzile F. Comparison of genetic diversity between Canadian adapted genotypes and exotic germplasm of soybean. Genome, 2010,53(5):337-345. |
| [11] | Doyle J J. Isolation of plant DNA from fresh tissue. Focus, 1990,12(1):13-15. |
| [12] | Burnham K D, Francis D M, Dorrance A E, et al. Genetic diversity patterns among phytophthora resistant soybean plant introductions based on SSR markers. Crop Science, 2002,42(2):338-343. |
| [13] | Zhang J, Hu K, Li K J, et al. Simulating the effects of long-term discontinuous and continuous fertilization with straw return on crop yields and soil organic carbon dynamics using the DNDC model. Soil and Tillage Research, 2017,165:302-314. |
| [14] | 朱振东, 王化波, 王晓鸣, 等. 黑龙江主要栽培大豆品种(系)对大豆疫霉根腐病的多抗性评价. 植物遗传资源学报, 2004,5(1):22-25. |
| [15] | 王衍莉, 杨义明, 范书田, 等. 基于SSR分子标记的73份山葡萄及杂交后代的遗传多样性分析. 生物技术通报, 2021,37(1):189-197. |
| [16] | 石慧, 王思明. 大豆在中国的历史变迁及其动因探究. 农业考古, 2019(3):32-39. |
| [17] | 文自翔. 中国栽培和野生大豆的遗传多样性、群体分化和演化及其育种性状QTL的关联分析. 南京:南京农业大学, 2008. |
| [18] | 朱申龙, Morti M L, Rao R. 应用AFLP方法研究中国大豆的遗传多样性. 浙江农业学报, 1998,10(6):302-309. |
| [19] | 周新安, 彭玉华, 王国勋, 等. 中国栽培大豆遗传多样性和起源中心初探. 中国农业科学, 1998,31(3):37-43. |
| [20] | 王彩洁, 孙石, 金素娟, 等. 中国大豆主产区不同年代大面积种植品种的遗传多样性分析. 作物学报, 2013,39(11):1917-1926. |
| [21] | 盖钧镒. 作物遗传育种学各论:第2版. 北京: 中国农业出版社, 1997. |
| [22] | 宋喜娥, 李英慧, 常汝镇, 等. 中国栽培大豆[Glycine max (L.) Merr.]微核心种质的群体结构与遗传多样性. 中国农业科学, 2010,43(11):2209-2219. |
| [23] | Ude G N, William J, Kenworthy , et al. Genetic diversity of soybean cultivars from China,Japan,North America,and North American ancestral lines determined by amplified fragment length polymorphism. Crop Science, 2003,43(5):1858-1867. |
| [24] | Nichols D M, Wang L Z, Yan L P, et al. Variability among Chinese Glycine soja and Chinese and North American soybean genotypes. Crop Science, 2007,47(3):1289-1298. |
| [25] | 盖钧镒, 赵团结. 中国大豆育种的核心祖先亲本分析. 南京农业大学学报, 2001,24(2):20-23. |
| [26] | 田清震, 盖钧镒, 喻德跃, 等. 我国野生大豆与栽培大豆AFLP指纹图谱研究. 中国农业科学, 2001,34(5):480-485. |
| [27] | 盖钧镒. 植物种质群体遗传结构改变的测度. 植物遗传资源学报, 2005,6(1):1-8,14. |
| [28] | 韩天富, 盖钧镒. 世界菜用大豆生产、贸易和研究的进展. 大豆科学, 2002,21(4):278-284. |
| [1] | Zhang Quanfang, Jiang Mingsong, Chen Feng, Zhu Wenyin, Zhou Xuebiao, Yang Lianqun, Xu Jiandi. Analysis of Genetic Diversity of Rice Varieties (Lines) in Shandong Province [J]. Crops, 2021, 37(4): 26-31. |
| [2] | Jia Ruiling, Zhao Xiaoqin, Nan Ming, Chen Fu, Liu Yanming, Wei Liping, Liu Junxiu, Ma Ning. Genetic Diversity Analysis and Comprehensive Assessment of Agronomic Traits of 64 Tartary Buckwheat Germplasms [J]. Crops, 2021, 37(3): 19-27. |
| [3] | Chu Guanghong, Zhang Jianxin, Wang Cong, Zhao Zhanying. Effects of Topping at Different Nodes at Seedling Stage on Root Growth and Yield of High-Yield Spring Soybean [J]. Crops, 2021, 37(3): 195-201. |
| [4] | Qin Ning, Li Junru, Li Wenlong, Du Hui, Li Xihuan, Zhang Caiying. Screening of Elite Germplasms and Identification of Seed Tocopherol and Its Component Contents in Soybean [J]. Crops, 2021, 37(3): 34-39. |
| [5] | Xiang Chao, Sun Suli, Zhu Zhendong, Zong Xuxiao, Yang Tao, Liu Rong, Yang Mei, Xian Dongfeng, Yang Xiuyan. Resistance and Molecular Identification to Powdery Mildew of Pea Germplasms in Sichuan [J]. Crops, 2021, 37(3): 51-56. |
| [6] | Su Yang, Yang Jing, Guo Yong, Du Weijun, Qiu Lijuan. Whole Genome Discovery and Analysis of Genes Related to 100-Seed Weight in Soybean [J]. Crops, 2021, 37(3): 8-18. |
| [7] | Li Candong, Guo Tai, Wang Zhixin, Zheng Wei, Zhao Haihong, Zhang Zhenyu, Xu Jiefei, Guo Meiling. Evaluation and Determination of Yield Evaluation Indicators of Soybean Mainly Cultivated Varieties in the Central and Eastern of Heilongjiang Province [J]. Crops, 2021, 37(2): 45-51. |
| [8] | Ma Mingchuan, Liu Longlong, Liu Zhang, Zhou Jianping, Nan Chenghu, Zhang Lijun. Analysis of SSR Loci in Whole Genome and Development of Molecular Markers in Tartary Buckwheat [J]. Crops, 2021, 37(1): 38-46. |
| [9] | Pan Xiaoxue, Hu Mingyu, Wang Zhongwei, Wu Hong, Lei Kairong. Evaluation of Agronomic Traits and Cold Tolerance at Germination Stage in Rice (Oryza sativa L.) Germplasms [J]. Crops, 2021, 37(1): 47-53. |
| [10] | Yang Wanjun, Pan Xiangyu, Wang Xiuhua, Wang Lu, Zhao Yan. Genetic Diversity Analysis of Yield and Agronomic Traits of 119 Alfalfa Varieties (Lines) [J]. Crops, 2020, 36(6): 17-22. |
| [11] | Wang Caijin, Di Wenjing, Ma Shumei, Wang Yang. Mining the Elite Allele of Resistance of Cercospora sojina Hara Race 1 in Soybean Resources [J]. Crops, 2020, 36(6): 189-196. |
| [12] | Gao Jie, Feng Guangcai, Li Xiaorong, Li Qingfeng, Peng Qiu. Phenotypic Diversity and Clustering Analysis of Sorghum Germplasm Resources in Different Regions of Guizhou Province [J]. Crops, 2020, 36(6): 54-60. |
| [13] | Zhao Yuyang, Song Jian, Qiu Lijuan. Proteomic Comparation Analysis of Thylakoid in Leaves of G-Locus Near Isogenic Line in Soybean [J]. Crops, 2020, 36(6): 8-16. |
| [14] | Gong Yanlong, Lei Yue, Yan Zhiqiang, Liu Xuewei, Zhang Dashuang, Wu Jianqiang, Zhu Susong. Comprehensive Evaluation of Phenotype Genetic Diversity in Japonica Rice Germplasm Resources in Different Ecological Zones [J]. Crops, 2020, 36(5): 71-79. |
| [15] | Chen Weiguo, Zhang Zheng, Shi Yugang, Cao Yaping, Wang Shuguang, Li Hong, Sun Daizhen. Drought-Tolerance Evaluation of 211 Wheat Germplasm Resources [J]. Crops, 2020, 36(4): 53-63. |
|
||