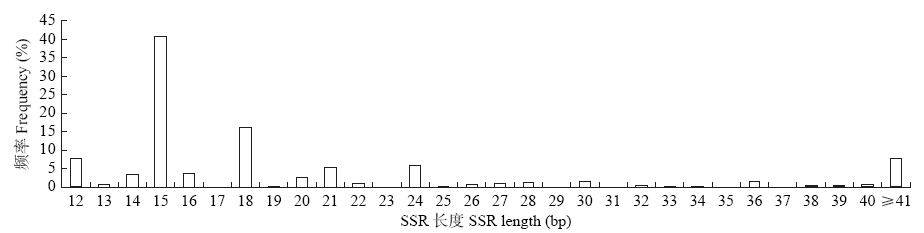
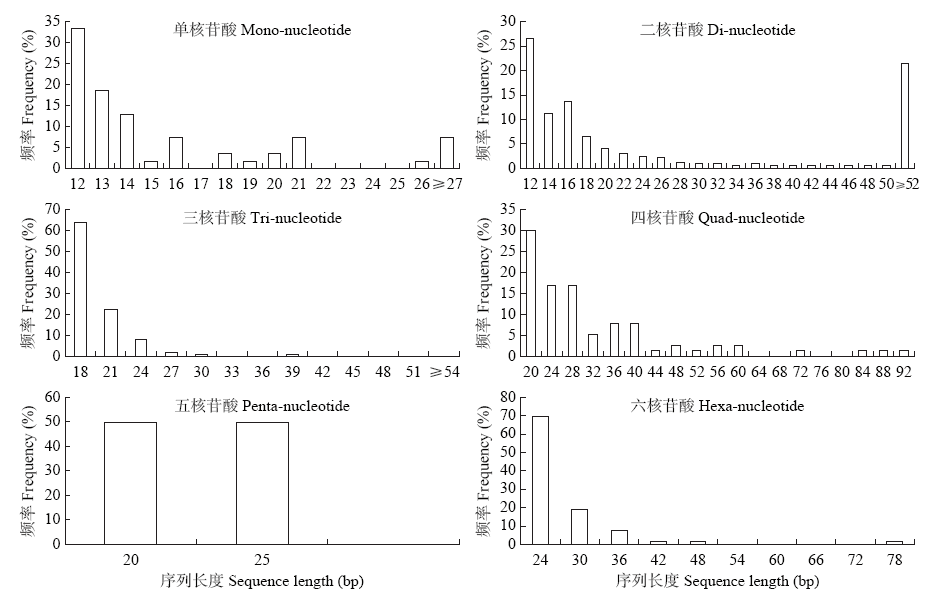
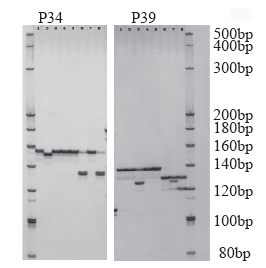
Crops ›› 2021, Vol. 37 ›› Issue (1): 38-46.doi: 10.16035/j.issn.1001-7283.2021.01.006
Previous Articles Next Articles
Analysis of SSR Loci in Whole Genome and Development of Molecular Markers in Tartary Buckwheat
Ma Mingchuan( ), Liu Longlong, Liu Zhang, Zhou Jianping, Nan Chenghu, Zhang Lijun(
), Liu Longlong, Liu Zhang, Zhou Jianping, Nan Chenghu, Zhang Lijun( )
)
- Center for Agricultural Genetic Resources Research, Shanxi Agricultural University/Key Laboratory of Crop Gene Resources and Germplasm Enhancement on Loess Plateau, Ministry of Agriculture and Rural Affairs/Shanxi Key Laboratory of Genetic Resources and Genetic Improvement of Minor Crops, Taiyuan 030031, Shanxi, China
| [1] | 胡银岗, 冯佰利, 周济铭, 等. 荞麦遗传资源利用及其改良研究进展. 西北农业学报, 2005,14(5):101-109. |
| [2] | 张丽君, 马名川, 刘龙龙, 等. 山西省苦荞品种资源研究. 河北农业科学, 2015,19(1):69-74. |
| [3] | 李秀莲, 史兴海, 朱慧珺. 苦荞产品开发应用现状及发展对策. 山西农业科学, 2011,39(8):908-910. |
| [4] | 王玲玲, 陈东亮, 黄丛林, 等. SSR分子标记技术在植物研究中的应用. 安徽农业科学, 2017,45(36)::123,126,130. |
| [5] | 杨梦婷, 黄洲, 干建平, 等. SSR分子标记的研究进展. 杭州师范大学学报(自然科学版), 2019,18(4):429-436. |
| [6] |
Iwata H, Imon K, Tsumura Y, et al. Genetic diversity among Japanese indigenous common buckwheat (Fagopyrum esculentum) cultivars as determined from amplified fragment length polymorphism and simple sequence repeat markers and quantitative agronomic traits. Genome, 2005,48(3):367-377.
doi: 10.1139/g04-121 pmid: 16121234 |
| [7] |
Konishi T, Yasui Y, Ohnishi O. Original birthplace of cultivated common buckwheat inferred from genetic relationships among cultivated populations and natural populations of wild common buckwheat revealed by AFLP analysis. Genes and Genetic Systems, 2005,80(2):113-119.
doi: 10.1266/ggs.80.113 pmid: 16172523 |
| [8] | 候雅君, 张宗文, 吴斌. 苦荞种质资源AFLP标记遗传多样性分析. 中国农业科学, 2009,42(12):4166-4174. |
| [9] | 王莉花, 殷富有, 刘继梅. 利用RAPD分析云南野生荞麦资源的多样性和亲缘关系. 分子植物育种, 2004,2(6):807-815. |
| [10] |
Tsuji K, Ohnishi O. Origin of cultivated tartary buckwheat (Fagopyrum tataricum Gaertn.) revealed by RAPD analyses. Genetic Resources and Crop Evolution, 2000,47:431-438.
doi: 10.1023/A:1008741513277 |
| [11] |
Kump B, Javornik B. Genetic diversity and relationships among cultivated and wild accessions of tartary buckwheat (Fagopyrum tataricum Gaertn.) as revealed by RAPD markers. Genetic Resources and Crop Evolution, 2002,49(6):565-572.
doi: 10.1023/A:1021250300572 |
| [12] | 邓琳琼, 张奎, 黄凯丰, 等. 甜荞和苦荞品种遗传多样性的RAPD分析. 安徽农业科学, 2011,39(15):8895-8898. |
| [13] |
Li Y Q, Fan X L, Shi T L, et al. SRAP marker reveals genetic diversity in tartary buckwheat in China. Frontiers of Agriculture in China, 2009,3(4):383-387.
doi: 10.1007/s11703-009-0071-5 |
| [14] |
Konishi T, Iwata H, Yashiro K, et al. Development and characterization of microsatellite markers for common buckwheat. Breed Science, 2006,56:277-285.
doi: 10.1270/jsbbs.56.277 |
| [15] |
Ma K H, Kim N S, Lee G A, et al. Development of SSR markers for studies of diversity in the genus Fagopyrum. Theoretical and Applied Genetics, 2009,119:1247-1254.
doi: 10.1007/s00122-009-1129-8 pmid: 19680622 |
| [16] |
Li Y Q, Shi T L, Zhang Z W. Development of microsatellite markers from tartary buckwheat. Biotechnology Letters, 2007,29(5):823-827.
doi: 10.1007/s10529-006-9293-2 pmid: 17216537 |
| [17] |
高帆, 张宗文, 吴斌. 中国苦荞SSR分子标记体系构建及其在遗传多样性分析中的应用. 中国农业科学, 2012,45(6):1042-1053.
doi: 10.3864/j.issn.0578-1752.2012.06.002 |
| [18] | 韩瑞霞, 张宗文, 吴斌. 苦荞SSR引物开发及其在遗传多样性分析中的应用. 植物遗传资源学报, 2012,13(5):759-764. |
| [19] | 石桃雄, 黎瑞源, 郭菊卉, 等. 基于普通荞麦种子表达序列标签微卫星标记的开发. 贵州农业科学, 2014,42(3):1-5. |
| [20] | 黎瑞源, 潘凡, 陈庆富, 等. 苦荞转录组EST-SSR发掘及多态性分析. 中国农业科技导报, 2015,17(4):42-52. |
| [21] | 杜晓磊, 张宗文, 吴斌, 等. 苦荞SSR分子遗传图谱的构建及分析. 中国农学通报, 2013,29(21):61-65. |
| [22] | 黎瑞源, 梁龙兵, 石桃雄, 等. 苦荞重组自交系群体F5代SSR遗传图谱的构建. 贵州师范大学学报(自然科学版), 2017,35(4):31-45. |
| [23] | 莫日更朝格图, 王鹏科, 高金锋, 等. 苦荞地方种质资源的遗传多样性分析. 西北植物学报, 2010,30(2):255-261. |
| [24] | 田晓庆, 徐宏亚, 汪灿, 等. 用SSR标记分析荞麦栽培种质资源的遗传多样性. 作物杂志, 2013(5):28-32. |
| [25] | 杨学文, 丁素荣, 胡陶, 等. 104份苦荞种质的遗传多样性分析. 作物杂志, 2013(6):13-17. |
| [26] | 徐笑宇, 方正武, 杨璞, 等. 苦荞遗传多样性分析与核心种质筛选. 干旱地区农业研究, 2015,33(1):268-277. |
| [27] |
Zhang L, Li X, Ma B, et al. The tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Molecular Plant, 2017,10(9):1224-1237.
pmid: 28866080 |
| [28] |
Varshney R K, Graner A, Sorrells M E. Genic microsatellite markers in plants:features and applications. Trends in Biotechnology, 2005,23(1):48-55.
pmid: 15629858 |
| [29] | Lawson M J, Zhang L Q. Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes. Genome Biology, 2006,7(2):R14.1-R14.11. |
| [30] | 原志敏. 玉米全基因组 SSRs 分子标记开发与特征分析. 雅安:四川农业大学, 2013. |
| [31] |
Kantety R V, Rota M L, Matthews D E, et al. Data mining for simple sequence repeats in expressed sequence tags from barley,maize,rice,sorghum and wheat. Plant Molecular Biology, 2002,48:501-510.
pmid: 11999831 |
| [32] |
王玉龙, 黄冰艳, 王思雨, 等. 四倍体野生种花生A. monticola全基因组SSR的开发与特征分析. 中国农业科学, 2019,52(15):2567-2580.
doi: 10.3864/j.issn.0578-1752.2019.15.002 |
| [33] | 李海洋, 李荣华, 夏岩石, 等. 基于RAD-seq数据开发烟草多态性SSR标记. 中国烟草科学, 2018,39(1):1-9. |
| [34] | Song Q, Jia G, Zhu Y, et al. Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR1.0) in soybean. Crop Science, 2010,50(5):1950-1960. |
| [35] | 郑燕, 张耿, 吴为人. 禾本科植物微卫星序列的特征分析和比较. 基因组学与应用生物学, 2011,30(5):513-520. |
| [36] |
Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nature Genetics, 2002,30(2):194-200.
doi: 10.1038/ng822 pmid: 11799393 |
| [37] | Dreisigacker S, Zhang P, Warburton M L, et al. SSR and pedigree analyses of genetic diversity among CIMMYT wheat lines targeted to different mega environments. Crop Science, 2004,44(2):381-388. |
| [38] |
Temnykh S, DeClerck G, Lukashova A, et al. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.):Frequency,length variation,transposon associations,and genetic marker potential. Genome Research, 2001,11(8):1441-1452.
doi: 10.1101/gr.184001 pmid: 11483586 |
| [39] |
陈仕勇, 马啸, 张新全, 等. 不同来源SSR和EST-SSR在披碱草属和鹅观草属物种中的通用性分析. 草业学报, 2016,25(2):132-140.
doi: 10.11686/cyxb2015324 |
| [40] | 卢玉飞, 蒋建雄, 易自力. 玉米SSR引物和甘蔗EST-SSR引物在芒属中的通用性研究. 草业学报, 2012,21(5):674-679. |
| [1] | Jin Jiangang, Tian Zaifang. Grey Correlation Analysis of Introduced Tartary Buckwheat in the Northern Shanxi [J]. Crops, 2021, 37(2): 52-56. |
| [2] | Lu Xiaoling, He Ming, Zhang Kaixuan, Liao Zhiyong, Zhou Meiliang. Study on the Cloning and Transformation of Rhamnose Transferase FtF3GT1 Gene in Tartary Buckwheat [J]. Crops, 2020, 36(5): 33-40. |
| [3] | Yang Xuele, Zhang Lu, Li Zhiqing, He Luqiu. Diversity Analysis of Tartary Buckwheat Germplasms Based on Phenotypic Traits [J]. Crops, 2020, 36(5): 53-58. |
| [4] | Gong Dan, Wang Suhua, Cheng Xuzhen, Wang Lixia. Construction of SSR Fingerprints and Diversity Analysis of a Cowpea Applied Core Collection [J]. Crops, 2020, 36(4): 79-83. |
| [5] | Li Chunhua, Huang Jinliang, Yin Guifang, Wang Yanqing, Lu Wenjie, Sun Daowang, Wang Chunlong, Guo Laichun, Hong Bo, Ren Changzhong, Wang Lihua. Genetic Analysis of Grain Shape Related Traits in Tartary Buckwheat [J]. Crops, 2020, 36(3): 42-46. |
| [6] | Li Hongqin, Liu Baolong, Zhang Bo, Zhang Huaigang. Analysis of Genetic Diversity and Establishment of Molecular ID of the Wheat Cultivars Registered in Qinghai Using SSR [J]. Crops, 2020, 36(3): 60-65. |
| [7] | Ma Mengli,Wang Tiantao,Lei En,Meng Hengling,Zhang Wei,Zhang Tingting,Lu Bingyue. Genetic Diversity Analysis of Amomum tsao-ko in Jinping Based on Phenotypic Traits and SSR Markers [J]. Crops, 2020, 36(2): 54-59. |
| [8] | Chengrui Ma,Dabing Xiang,Yan Wan,Jianyong Ouyang,Yue Song,Zhengsong Tang,Jianying Liu,Gang Zhao. Difference Analysis of Spatial Distribution Characteristics of Different Tartary Buckwheat Varieties [J]. Crops, 2020, 36(1): 35-40. |
| [9] | Yang Tian,Zhang Yongqing,Dong Fuhui,Ma Xingxing,Xue Xiaojiao. Research on the Root Growth of Different Drought-Resistant Fagopyrum tataricum under Different Water Conditions [J]. Crops, 2019, 35(6): 76-82. |
| [10] | Li Song,Zhang Shicheng,Dong Yunwu,Shi Delin,Shi Yundong. Genetic Diversity Analysis of Rice Varieties in Tengchong, Yunnan Based on SSR Markers [J]. Crops, 2019, 35(5): 15-21. |
| [11] | Song Lifang,Feng Meichen,Zhang Meijun,Xiao Lujie,Wang Chao,Yang Wude,Song Xiaoyan. Effects of Exogenous Selenium on the Growth and Development of Tartary Buckwheat and Selenium Content in Grains [J]. Crops, 2019, 35(3): 150-154. |
| [12] | Ma Mingchuan,Liu Longlong,Zhang Lijun,Cui Lin,Zhou Jianping. Morphological Identification and Analysis of EMS-Induced Mutants from Ciqiao [J]. Crops, 2019, 35(3): 37-41. |
| [13] | Yasong Cui, Yan Wang, Lijuan Yang, Chaoxin Wu, Piao Zhou, Pan Ran, Qingfu Chen. Genetic Analysis of Fruit Hull Rate and Related Traits on Tartary Buckwheat [J]. Crops, 2019, 35(2): 51-60. |
| [14] | Lei Wu,Lanfen Wang,Jing Wu,Shumin Wang. Association Analysis of Root-Related Traits in Common Bean [J]. Crops, 2019, 35(2): 61-70. |
| [15] | Chunyu Lin,Xiaoyu Liang,Huiyan Zhao,Yang Wang. Analysis of Genetic Diversity and Population Structure of Main Soybean Varieties in Heilongjiang Province [J]. Crops, 2019, 35(2): 78-83. |
|