Crops ›› 2022, Vol. 38 ›› Issue (6): 42-53.doi: 10.16035/j.issn.1001-7283.2022.06.006
Previous Articles Next Articles
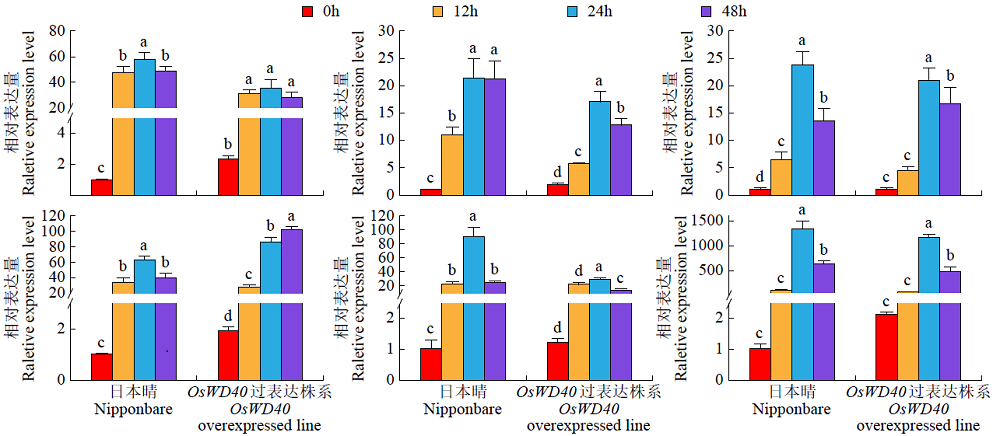
Transcriptome Analysis of OsWD40 Overexpression Rice Roots in Response to Salt Stress
Wen Danni( ), Bao Lingran, Liu Mengmeng, Shen Bo(
), Bao Lingran, Liu Mengmeng, Shen Bo( )
)
- College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou 311121, Zhejiang, China
| [1] |
Almeida D M, Oliveira M M, Saibo N J M. Regulation of Na+ and K+ homeostasis in plants:towards improved salt stress tolerance in crop plants. Genetics and Molecular Biology, 2017, 40(1):326-345.
doi: 10.1590/1678-4685-gmb-2016-0106 |
| [2] |
Senadheera P, Singh R, Maathuis F J. Differentially expressed membrane transporters in rice roots may contribute to cultivar dependent salt tolerance. Journal of Experimental Botany, 2009, 60(9):2553-2563.
doi: 10.1093/jxb/erp099 pmid: 19395386 |
| [3] |
Negrao S, Schmockel S M, Tester M, et al. Evaluating physiological responses of plants to salinity stress. Annals of Botany, 2017, 119:1-11.
doi: 10.1093/aob/mcw191 pmid: 27707746 |
| [4] |
梁永书, 周军杰, 南文斌, 等. 水稻根系研究进展. 植物学报, 2016, 51(1):98-106.
doi: 10.11983/CBB15011 |
| [5] |
Stirnimann C U, Petsalaki E, Russell R B, et al. WD40 proteins propel cellular networks. Trends in Biochemical Sciences, 2010, 35(10):565-574.
doi: 10.1016/j.tibs.2010.04.003 pmid: 20451393 |
| [6] |
Smith T F. Diversity of WD-repeat proteins. Sub-Cellular Biochemistry, 2008, 48:20-30.
doi: 10.1007/978-0-387-09595-0_3 pmid: 18925368 |
| [7] |
Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell, 2011, 2(3):202-214.
doi: 10.1007/s13238-011-1018-1 pmid: 21468892 |
| [8] |
Smith T F, Gaitatzes C, Saxena K, et al. The WD repeat:a common architecture for diverse functions. Trends in Biochemical Sciences, 1999, 24(5):181-185.
pmid: 10322433 |
| [9] | Nocker S V, Ludwig P. The WD-repeat protein superfamily in Arabidopsis:conservation and divergence in structure and function. BMC Genomics, 2003, 4(1):297-300. |
| [10] |
Xu X Z, Wan W, Jiang G B, et al. Nucleocytoplasmic trafficking of the Arabidopsis WD 40 repeat protein XIW1 regulates ABI5 stability and abscisic acid responses. Molecular Plant, 2019, 12(12):1598-1611.
doi: 10.1016/j.molp.2019.07.001 |
| [11] |
Kong D J, Li M J, Dong Z H, et al. Identification of TaWD40D,a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Reports, 2015, 34:395-410.
doi: 10.1007/s00299-014-1717-1 |
| [12] |
Huang J, Wang M M, Bao Y M, et al. SRWD:A novel WD 40 protein subfamily regulated by salt stress in rice (Oryza sativa L.). Gene, 2008, 424(1):71-79.
doi: 10.1016/j.gene.2008.07.027 |
| [13] |
Wang J, Zhang Y D, Fan F J, et al. Comparative transcriptome analysis reveals molecular response to salinity stress of salt-tolerant and sensitive genotypes of indica rice at seedling stage. Scientific Reports, 2018, 8(1):1755-1767.
doi: 10.1038/s41598-018-20191-w |
| [14] |
Chen E Y, Shen B. OsABT,a rice WD40 domain-containing protein,is involved in abiotic stress tolerance. Rice Science, 2022, 29(3):247-256.
doi: 10.1016/j.rsci.2021.07.012 |
| [15] |
Wang A J, Shu X Y, Jing X, et al. Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnology Journal, 2021, 19(8):1553-1566.
doi: 10.1111/pbi.13569 |
| [16] |
Horie T, Sugawara M, Okada T, et al. Rice sodium-insensitive potassium transporter,OsHAK5,confers increased salt tolerance in tobacco BY2 cells. Journal of Bioscience and Bioengineering, 2011, 111(3):346-356.
doi: 10.1016/j.jbiosc.2010.10.014 |
| [17] |
Yang T, Feng H, Zhang S, et al. The potassium transporter OsHAK 5 alters rice architecture via ATP-dependent transmembrane auxin fluxes. Plant Communications, 2020, 1(5):100052.
doi: 10.1016/j.xplc.2020.100052 |
| [18] |
Sah S K, Reddy K R, Li J X. Abscisic acid and abiotic stress tolerance in crop plants. Frontiers in Plant Science, 2016, 7:571.
doi: 10.3389/fpls.2016.00571 pmid: 27200044 |
| [19] |
Nambara E, Marion P A. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology, 2005, 56:165-185.
pmid: 15862093 |
| [20] |
Kim H, Lee K, Hwang H, et al. Overexpression of PYL5 in rice enhances drought tolerance,inhibits growth,and modulates gene expression. Journal of Experimental Botany, 2014, 65(2):453-464.
doi: 10.1093/jxb/ert397 |
| [21] |
Zhou M J, Guan Y H, Ren H B, et al. A bZIP transcription factor,OsABI5,is involved in rice fertility and stress tolerance. Plant Molecular Biology, 2008, 66(6):675-683.
doi: 10.1007/s11103-008-9298-4 |
| [22] | 孙立影, 于志晶, 李海云. 植物次生代谢物研究进展. 吉林农业科学, 2009, 34(4):4-10. |
| [23] |
Fraser C M, Chaoole C. The Phenylpropanoid Pathway in Arabidopsis. The Arabidopsis Book, 2011, 9:e0152.
doi: 10.1199/tab.0152 |
| [24] |
Gui J S, Shen J H, Li L G. Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiology, 2011, 157(2):574-586.
doi: 10.1104/pp.111.178301 pmid: 21807887 |
| [25] | 崔慧萍, 周薇, 郭长虹. 植物过氧化物酶体在活性氧信号网络中的作用. 中国生物化学与分子生物学报, 2017, 33(3):220-226. |
| [26] |
Huang J, Sun S, Xu D, et al. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Molecular Biology, 2012, 80(3):337-350.
doi: 10.1007/s11103-012-9955-5 pmid: 22930448 |
| [27] |
Devaiah B N, Karthikeyan A S, Raghothama K G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiology, 2007, 143(4):1789-1801.
doi: 10.1104/pp.106.093971 |
| [28] | Pillai S E, Kumar C, Patel H K, et al. Overexpression of a cell wall damage induced transcription factor,OsWRKY42,leads to enhanced callose deposition and tolerance to salt stress but does not enhance tolerance to bacterial infection. BioMed Central, 2018, 18(1):177. |
| [29] | Zhao Y, Xing L, Wang X A, et al. The ABA receptor PYL 8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Science Signaling, 2014, 7(328):ra53. |
| [30] |
Dai X Y, Xu Y Y, Ma Q B, et al. Overexpression of an R1R2R3 MYB gene,OsMYB3R-2,increases tolerance to freezing,drought,and salt stress in transgenic Arabidopsis. Plant Physiology, 2007, 143(4):1739-1751.
doi: 10.1104/pp.106.094532 |
| [31] |
Liu G Z, Li X L, Jin S X, et al. Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS ONE, 2017, 9(1):e86895.
doi: 10.1371/journal.pone.0086895 |
| [32] |
Schiefelbein J, Huang L, Zheng X H. Regulation of epidermal cell fate in Arabidopsis roots:the importance of multiple feedback loops. Frontiers in Plant Science, 2014, 5:47.
doi: 10.3389/fpls.2014.00047 pmid: 24596575 |
| [33] |
Li J Z, Han Y C, Lei L, et al. qRT9,a quantitative trait locus controlling root thickness and root length in upland rice. Journal of Experimental Botany, 2015, 66(9):2723-2732.
doi: 10.1093/jxb/erv076 |
| [34] |
Cao Y F, Song F M, Goodman R M, et al. Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. Journal of Plant Physiology, 2006, 163(11):1167-1178.
pmid: 16436304 |
| [1] | Zhou Hao, Qiu Xianjin, Xu Jianlong. Advance in Effects of Magnetized Water Irrigation on Crop Growth and Development [J]. Crops, 2022, 38(6): 1-6. |
| [2] | Xiong Yousheng, Xiong Hanfeng, Guo Yanlong, Wang Haisheng, Liu Wei, Yan Yuxiang, Xie Yuanyuan, Zhou Jianxiong, Yang Lijun. Effects of Reducing Fertilizer Application Models on Wheat Yield and Nutrient Use Efficiencies in Rice-Wheat Cropping System [J]. Crops, 2022, 38(6): 118-123. |
| [3] | Qin Meng, Cui Shize, He Xiaodong, Zhai Lingxia, Tao Bo, Wang Zhaojun, Zhao Haicheng, Li Hongyu, Zheng Guiping, Liu Lihua. Effects of Straw Puffing Returning on Rice Yield, Quality and Soil Nutrients [J]. Crops, 2022, 38(6): 159-166. |
| [4] | Jiang Shukun, Wang Lizhi, Yang Xianli, Zhang Xijuan, Liu Kai, Chi Liyong, Li Rui, Lai Yongcai. Spatiotemporal Change Characteristics of Rice Growth Climate Resources in Saline-Alkaline Area of Songnen Plain from 1961 to 2019 [J]. Crops, 2022, 38(6): 214-219. |
| [5] | Chong Haotian, Shang Cheng, Zhang Yunbo, Huang Liying. Effects of Dense Planting with Reduced Nitrogen Application on Spikelet Formation of Different Types of Rice Varieties [J]. Crops, 2022, 38(6): 226-233. |
| [6] | Wang Jinxiang, Wang Yanzhi, Xing Lixuan, Liu Jianxia, Wang Runmei. Effects of GA3 on Root Growth and Osmotic Regulation of Lübaonuo Broomcorn Millet Seedlings under Salt Stress [J]. Crops, 2022, 38(6): 98-104. |
| [7] | Wang Hanxiang, Li Guangcun, Xu Jianfei, Wang Wanxing, Jin Liping. Advances in Research on Salt Tolerance Mechanism of Plants [J]. Crops, 2022, 38(5): 1-12. |
| [8] | Wang Yan, Li Tingyou, Wang Dou, Li Jiawei, Peng Wenlu, Rui Haiyun. Effects of Isosteviol on Growth of Wheat Seedlings under Salt Stress [J]. Crops, 2022, 38(5): 141-145. |
| [9] | Zhang Chonghua, Duan Licheng, Wang Shangming, Zhang Qingxia, Wang Chengzi, Wu Fengyu, Yang Lin. Effects of Sowing Date on Late-Rice Yield and Utilization of Heat-Light Resources in Jiangxi Province [J]. Crops, 2022, 38(5): 229-234. |
| [10] | Pan Junfeng, Liu Yanzhuo, Liang Kaiming, Huang Nongrong, Peng Bilin, Fu Youqiang, Hu Xiangyu, Zhong Xuhua, Li Meijuan, Hu Rui. Effects of Long- and Short-Term Reduction of Phosphorus Input on Yield and Phosphorus Utilization of Double Cropping Rice in South China [J]. Crops, 2022, 38(5): 241-248. |
| [11] | Li Rui, Dong Liqiang, Shang Wenqi, Yu Guangxing, Dai Guijin, Wang Zheng, Li Yuedong. Effects of Water Spraying Interval at Seedling Stage on Growth and Yield of Rice [J]. Crops, 2022, 38(5): 249-254. |
| [12] | Dong Linlin, Shen Mingxing, Shi Linlin, Shen Yuan, Wang Haihou, Lu Changying. The Effects of Biochar Combined with Earthworm Cast Application on Rice Yield and Nutrient Uptake [J]. Crops, 2022, 38(5): 69-77. |
| [13] | Zhou Yujiao, Zhang Weiyang, Yang Jianchang. Research Advances on High Temperature Induced-Impairment in Spikelet-Opening and Pistil-Fertilization of Photo-Thermo-Sensitive Genic Male Sterile Rice Lines [J]. Crops, 2022, 38(4): 1-8. |
| [14] | Chen Shiyong, Wang Rui, Chen Zhiqing, Zhang Haipeng, Wang Juanjuan, Shan Yuhua, Yang Yanju. Effects of Nano-Zinc and Ion-Zinc on Rice Yield Formation and Grain Zinc Content [J]. Crops, 2022, 38(4): 107-114. |
| [15] | Tang Jianpeng, Chen Jingdu, Wen Kai, Zhang Mingwei, Xie Chenglin, Lu Peiling, Min Sigui, Wang Qiluan, Cheng Jiemin. Study on Material Production and Yield Characteristics of Japonica Rice with Good Eating Quality in Rice-Crayfish Farming System [J]. Crops, 2022, 38(4): 115-123. |
|
||