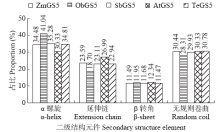
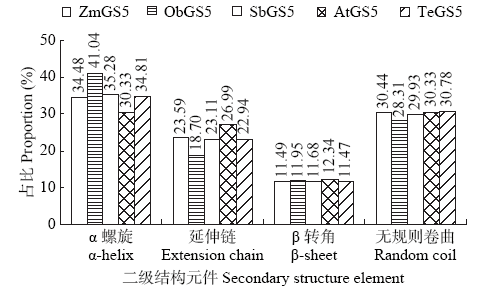
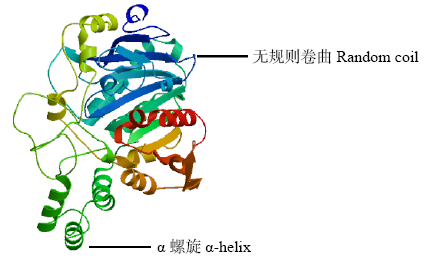
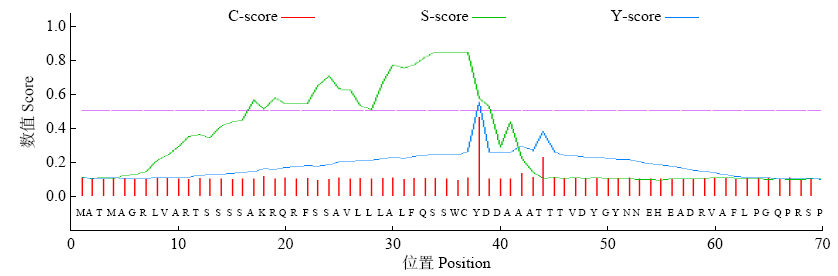
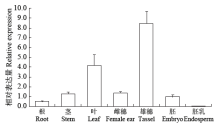
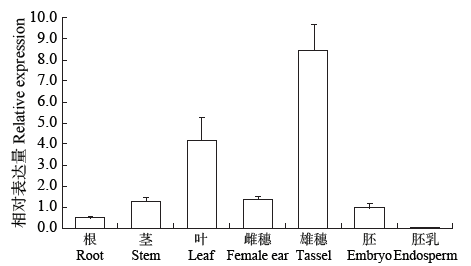
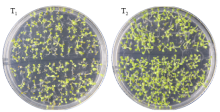
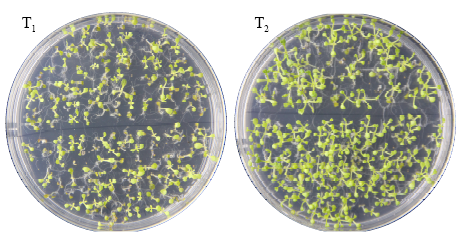
Crops ›› 2021, Vol. 37 ›› Issue (1): 16-25.doi: 10.16035/j.issn.1001-7283.2021.01.003
Previous Articles Next Articles
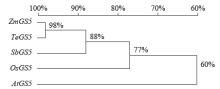
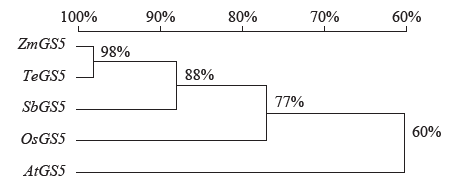
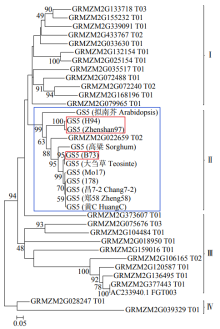
Cloning, Molecular Characteristics Analysis and Genetic Transformation of Arabidopsis thaliana of ZmGS5 Gene in Maize
Liu Xiaoli1,2( ), Han Litao1, Wei Nan1, Shen Fei1, Cai Yilin2(
), Han Litao1, Wei Nan1, Shen Fei1, Cai Yilin2( )
)
- 1Department of Food Engineering, Henan Vocational College of Agriculture, Zhengzhou 451450, Henan, China
2Maize Research Institute, Southwest University, Chongqing 400715, China
| [1] | 李向拓, 毛建昌, 吴权明. 分子标记在玉米育种中的应用. 玉米科学, 2004,43(2):26-29. |
| [2] | 贾波, 管飞翔, 谢庆春, 等. 玉米产量性状QTL定位分析. 西南农业学报, 2013,26(1):22-25. |
| [3] | 黄荣荣, 周子键, 陈甲法, 等. 玉米缺陷性籽粒突变体的遗传分析及突变基因dek1-T7的定位. 分子植物育种, 2012,10(2):163-168. |
| [4] |
Li X, Chen G H, Zhang W Y, et al. Genome-wide transcriptional analysis of maize endosperm in response to ae wx double mutations. Journal of Genetics and Genomics, 2010,37(11):749-762.
doi: 10.1016/S1673-8527(09)60092-8 pmid: 21115169 |
| [5] |
Yi G, Lauter A M, Scott M P, et al. The thick aleurone 1 mutant defines a negative regulation of maize aleurone cell fate that functions downstream of defective kernell. Plant Physiology, 2011,156:1826-1836.
doi: 10.1104/pp.111.177725 pmid: 21617032 |
| [6] | 宋同明, 陆效武. 对一个玉米双重标记新突变基因(os)的染色体定位和初步遗传研究. 遗传学报, 1993(5):432-438. |
| [7] | Lid S E, Gruis D, Jung R, et al. The defective kernel 1 (dek1) gene require for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proceedings of the National Academy of Sciences of the United States of America, 2002,8(99):5460-5465. |
| [8] |
Jahnke S, Scholten S. Epigenetic resetting of a gene imprinted in plant embryos. Current Biology, 2009,19:1677-1681.
doi: 10.1016/j.cub.2009.08.053 pmid: 19781944 |
| [9] |
Li Y, Fan C, Xing Y, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nature Genetics, 2011,43(12):1266-1269.
doi: 10.1038/ng.977 pmid: 22019783 |
| [10] |
Kellogg E A. Evolutionary history of the grasses. Plant Physiology, 2001,125(3):1198-1205.
pmid: 11244101 |
| [11] | 王建兵, 汤华, 黄益勤, 等. 玉米和水稻重要性状QTL的比较研究. 遗传学报, 2004,31(12):1401-1407. |
| [12] |
Raith M R, Kelty C A, Griffith J F, et al. Comparison of PCR and quantitative real-time PCR methods for the characterization of ruminant and cattle fecal pollution sources. Water Research, 2013,47(18):6921-6928.
doi: 10.1016/j.watres.2013.03.061 pmid: 23871256 |
| [13] | 刘晓丽, 魏楠. 拟南芥的遗传转化. 河南农业, 2019(27):45-46. |
| [14] | Xu C J, Liu Y, Li Y B, et al. Differential expression of GS5 regulates grain size in rice. Journal of Experimental Botany, 2015,9(66):2611-2623. |
| [15] | 胡甘雨, 司风铃, 车燕飞, 等. 葱蝇过氧化氢酶基因的克隆及生物信息学分析. 西南大学学报(自然科学版), 2014,36(2):32-40. |
| [16] |
Mahoney J A, Ntolosi B, DaSilva R P, et al. Cloning and characterization of CPVL,a novel serine carboxypeptidase,from human macrophages. Genomics, 2001,72:243-251.
doi: 10.1006/geno.2000.6484 pmid: 11401439 |
| [17] | Shirley A M, Chapple C. Biochemical characterization of sinapoyglucose choline sinapolytransferase,a serine carboxypeptidase-like protein that functions as an acyltransferase in plant secondary metabolism. Journal Biological Chemistry, 2003,278(22):19870-19877. |
| [18] |
Cercós M, Urbez C, Carbonell J. A serine carboxypeptidase gene (PsCP),expressed in early steps of reproductive and vegetative development in Pisum sativum,is induced by gibberellins. Plant Molecular Biology, 2003,51(2):165-174.
doi: 10.1023/a:1021142403856 pmid: 12602875 |
| [19] |
刘丽, 王静, 张志明, 等. 玉米丝氨酸羧肽酶基因(ZmSCP)的克隆及表达分析. 作物学报, 2013,39(1):164-171.
doi: 10.3724/SP.J.1006.2013.00164 |
| [20] |
Liu H Z, Wang X E, Zhang H J, et al. A rice serine carboxypeptidase-like gene OsBISCPL1 is involved in regulation of defense responses against biotic and oxidative stress. Gene, 2008,420:57-65.
pmid: 18571878 |
| [1] | Li Yanfang,Du Yanwei,Zhang Zheng,Wang Gaohong,Zhao Genyou,Zhao Jinfeng,Yu Aili. Establishment and Optimization of Agrobacterium Mediated Transformation System for Mature Embryo of Foxtail Millet [J]. Crops, 2019, 35(3): 73-79. |
| [2] | Haibin Luo, Shengli Jiang, Chengmei Huang, Huiqing Cao, Zhinian Deng, Kaichao Wu, Lin Xu, Zhen Lu, Yuanwen Wei. Cloning and Expression of ScHAK10 Gene in Sugarcane [J]. Crops, 2018, 34(4): 53-61. |
| [3] | Wei Zhang,Liangqun Wang,Yong Liu,Yanfang Hao,Wei Yang,Hongyan Bai,Bo Wu. Optimization of the Factors Related to the Efficiency of Agrobacterium-Mediated Transformation of Sorghum [J]. Crops, 2018, 34(1): 56-61. |
|