WRKY转录因子广泛参与植物系统发育[11-12]、逆境应答[13-14]和转录激活[15-16]等过程。LbWRKY3基因参与枸杞(Lycium barbarum L.)果实生长发育调控[17];CaWRKY40和CaWRKY6在辣椒(Capsicum annuum)应答青枯菌和高温胁迫的过程中具有正向调控作用[18];MeWRKY20、MeWRKY21和MeWRKY24等16个WRKY基因调控木薯(Manihot esculenta)抗细菌性病害响应[19]。近年来,随着大规模高通量测序的开展,越来越多的物种完成了WRKY家族信息的鉴定,如拟南芥包含72个WRKY成员[20],粳稻和籼稻分别有98和102个[21],番茄有81个[22],棉花有116个[23]等。
生姜是姜科植物姜(Zingiber officinale Roscoe)的根状茎,具有药食两用的特点,因具有单位面积产量高和经济效益好等巨大优势,是一种值得推广的经济作物[24]。然而,生姜在旺盛生长期(6-9月)易遭受环境水分和青枯菌侵染等胁迫,对其生长、品质和产量有重要的影响。借助生物信息学手段可深入分析生姜对逆境胁迫的响应,挖掘抗性基因,解析转录因子介导的抗性调控机制。由于生姜基因组杂合性较高的限制,基因组测序工作一直没有完成,导致分子生物学研究相对迟缓,特别是关于生姜ZoWRKY转录因子的研究更为少见。本研究在生姜转录组数据分析的基础上,鉴定了ZoWRKY家族成员,并分析了不同土壤湿度和青枯菌侵染下ZoWRKY基因的逆境响应模式,对后续开展ZoWRKY转录因子功能研究、推进生姜抗性育种和提高生姜产量具有重要意义。
1 材料与方法
1.1 试验材料
试验以生姜品种西南竹根姜(Zingiber officinale Roscoe cv. Zhugen)为材料。生姜组培苗栽培于重庆文理学院特色植物研究院温室。温度30℃,土壤湿度(充水孔隙度,water-filled pore spaces,WFPS)为10%、25%、30%和40%,光强200μE/(m2·s),光周期为14h光照/10h黑暗),生长至3股杈(约90d)时进行青枯菌(Ralstonia solanacearum)侵染处理。侵染接种前,对各湿度条件下的生姜根茎进行创伤,然后用浓度为106cfu/mL的青枯菌悬浮液对处理组生姜的土壤进行浸渍接种,同时,以无菌水接种为对照。生长至6个月,收集不同土壤湿度和青枯菌侵染处理的生姜地下肉质根、地下根茎、地上茎和叶片等组织,置于液氮中快速冷冻,-80℃中保存用于提取RNA。
1.2 RNA-Seq测序
按照Trizol试剂盒说明书分别提取生姜各组织总RNA,经检测合格后等量混合,基于Illumina公司Hiseq 4000(Illumina,美国)平台进行转录组测序,采用Trinity软件拼接获取Unigenes。
1.3 ZoWRKY转录因子基因筛选
利用Galaxy网站(
1.4 ZoWRKY转录因子的系统进化分析
从PlantTFDB数据库(
1.5 ZoWRKY序列保守结构域分析
利用DNAMAN 7.0软件分别对筛选到的ZoWRKY蛋白序列进行多序列比对,获得保守结构域区域。利用MEME(
1.6 ZoWRKY理化性质分析
采用ProtParam tool(
1.7 ZoWRKY基因的逆境响应模式分析
从NCBI网站下载不同WFPS(10%、25%、30%和40%)和青枯菌(R. solanacearum)侵染前后的生姜根茎转录组数据(Bioproject:PRJNA380972),利用Bowtie 2.0软件(
2 结果与分析
2.1 RNA-Seq测序及组装
用Illumina HiSeq™进行混合RNA样品二代转录组测序,共产生27 645 008对Paired-End reads(表1)。经过Trinity组装、cd-hit-est聚类后获得381 871条无冗余、长度大于300bp的Unigenes,平均长度为891bp,N50长度为1260bp,最大长度为16 953bp,GC比例为45.30%的340.2Mb生姜转录组数据。
表1 生姜转录组组装统计
Table 1
| 项目Item | 数值 Numerial number |
|---|---|
| 双末端读长数PE read number | 27 645 008 |
| 基因数Unigenes number | 381 871 |
| 长度≥10 000bp的基因 Unigenes≥10 000bp | 47 |
| 长度≥2000bp的基因 Unigenes≥2000bp | 19 182 |
| 长度≥1000bp的基因 Unigenes≥1000bp | 61 387 |
| 平均长度Average length (bp) | 891 |
| 最大长度Maximum length (bp) | 16 953 |
| N50长度N50 length (bp) | 1260 |
| 总长度Total length (bp) | 340 247 061 |
2.2 ZoWRKY筛选及全长序列的获取
381 871条Unigenes经EMBOSS程序预测获得106 653条蛋白序列(长度≥150),利用HMMER 3.0软件质询WRKY保守域种子序列(序列号PF03106),设置序列覆盖度>90%和Independent E value<0.01,共筛选获得78条序列不同的ZoWRKY蛋白。参照生姜近缘物种香蕉MaWRKY家族基因序列,其中72条ZoWRKY具有完整的ORF序列,而Zoff188265等6条仅有部分ORF序列。
对Zoff188265、Zoff210606、Zoff217771、Zoff244943、Zoff295642和Zoff614319等6条无完整ORF序列的ZoWRKY转录因子进行Race-PCR扩增。首先基于无完整ORF序列的ZoWRKY序列设计5′和3′端特异性引物,准备好反应所需酶系和体系,然后借助Race-PCR试剂盒(Clontech,Mountain View,美国)分别进行cDNA第1条链和第2条链的合成。Race-PCR试验成功获得其全长ORF序列,其中Zoff614319的ORF长度最短,为600bp,Zoff295642的ORF长度最长,为2499bp。
2.3 ZoWRKY系统进化分析
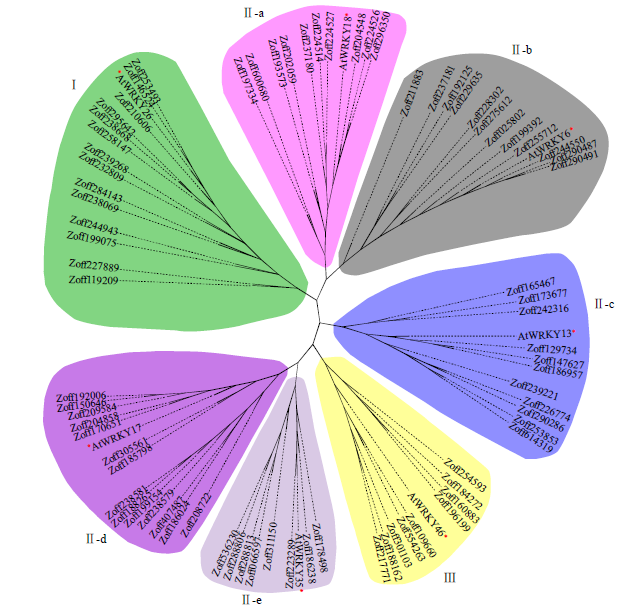
以拟南芥AtWRKY蛋白序列为指导,利用MEGA-X对AtWRKY和ZoWRKY蛋白序列进行多序列比对,用Neighbor Joining法构建系统进化树。根据7个不同亚族的AtWRKY序列与78条ZoWRKY序列的聚类情况,将ZoWRKY转录因子分成3个亚族,亚族Ⅰ有14个成员;亚族Ⅱ有55个成员,其中Ⅱ-a、Ⅱ-b、Ⅱ-c、Ⅱ-d和Ⅱ-e分别有10、12、11、14和8个成员;亚族Ⅲ有9个成员(图1)。
图1
图1
生姜与拟南芥WRKY转录因子家族成员系统进化关系
“*”表示拟南芥AtWRKY转录因子
Fig.1
Phylogenetic relationships of WRKY transcription factors in Zingiber and Arabidopsis
“*”represent the AtWRKY transcription factor
2.4 ZoWRKY保守结构域分析
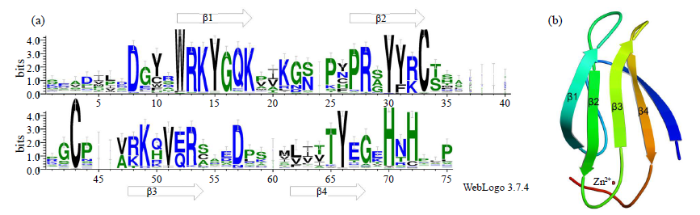
图2
图2
ZoWRKY蛋白保守结构域序列标签(a)及三维结构模型(b)
序列标签由每个位置的字母组成,字母相对大小表明它们在序列中的频率,字母总高度表示位置的信息内容,以位为单位
Fig.2
The sequence tags (a) and three-dimensional structural model (b) of the WRKY conserved domain in ZoWRKY proteins
A sequence tag consists of a stack of letters at each position, the relative sizes of the letters indicate their frequency in the sequences, the total height of the letters depicts the information content of the position, in bits
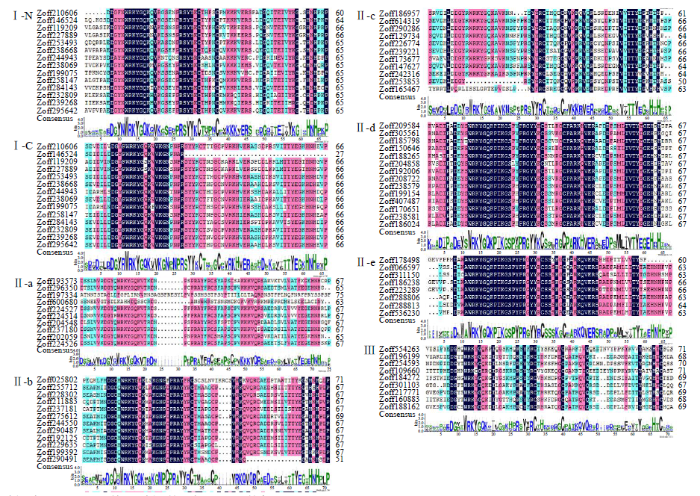
ZoWRKY亚族保守结构域分析结果(图3)显示,亚族成员的WRKY结构域保守度较高,个别成员发生WRKYGQK七肽域变异和锌指结构变异。亚族Ⅰ含有2个WRKY保守结构域,根据其在序列中的位置又进一步分为Ⅰ-N和Ⅰ-C亚族。亚族Ⅰ-N的WRKY七肽域为WRKYGQK,锌指结构为CX4CX22-23HXH形式。亚族Ⅰ-C的WRKY七肽域中为WRKYGQK,锌指结构为CX4CX23HXH形式。与亚族Ⅰ不同,亚族Ⅱ和Ⅲ只有1个WRKY保守结构域,根据亚族Ⅱ的氨基酸序列差异,进一步分为Ⅱ-a、Ⅱ-b、Ⅱ-c、Ⅱ-d和Ⅱ-e。其中,亚族Ⅱ-a、Ⅱ-b、Ⅱ-d和Ⅱ-e的WRKY七肽域和锌指结构为WRKYGQK和CX5CX23HXH形式,亚族Ⅱ-c的为WRKYGQK和CX4CX23HXH形式。亚族Ⅲ的WRKY七肽域和锌指结构为WRKYGQK和CX7CX23HXC形式。
图3
图3
ZoWRKY转录因子保守结构域分析
颜色表示序列保守性差异,颜色越深表示保守性越高
Fig.3
Sequence analysis of the WRKY conserved domain in ZoWRKY proteins
Colors indicate the sequence simility and a darker color had a higher conservatism
2.5 ZoWRKY氨基酸组成及结构预测分析
对ZoWRKY进行ProtParam分析(表2)表明,不同亚族之间的氨基酸残基数目、相对分子质量和等电点等理化性质存在差异。亚族Ⅰ的氨基酸残基数目最多,平均为490个,相对分子质量最高(53.6kDa);亚族Ⅱ-a的氨基酸残基数目最少,平均为210个,相对分子质量最低(23.5kDa)。ZoWRKY的等电点大多数在碱性范围。
表2 ZoWRKY转录因子氨基酸理化分析
Table 2
| 序号 Number | 序列ID Sequence ID | 类型 Type | 理论等电点 PI | 相对分子质量 MW (kDa) | 氨基酸长度 Amino acid length | α-螺旋 α-helix (%) | β-折叠 β-fold (%) | 其他 Others (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Zoff119209 | Ⅰ | 10.02 | 18 952.62 | 174 | 1.72 | 17.24 | 81.03 | ||||||||
| 2 | Zoff146524 | Ⅰ | 6.05 | 47 740.90 | 433 | 1.15 | 15.70 | 83.14 | ||||||||
| 3 | Zoff199075 | Ⅰ | 5.50 | 58 838.27 | 540 | 1.67 | 14.81 | 83.52 | ||||||||
| 4 | Zoff210606 | Ⅰ | 7.36 | 49 275.37 | 451 | 2.22 | 15.96 | 81.82 | ||||||||
| 5 | Zoff227889 | Ⅰ | 9.61 | 31 473.00 | 281 | 3.56 | 20.64 | 75.80 | ||||||||
| 6 | Zoff232809 | Ⅰ | 5.19 | 43 100.82 | 391 | 2.05 | 9.97 | 87.98 | ||||||||
| 7 | Zoff238069 | Ⅰ | 6.63 | 52 133.52 | 479 | 2.92 | 16.28 | 80.79 | ||||||||
| 8 | Zoff238668 | Ⅰ | 5.59 | 60 829.58 | 537 | 2.79 | 16.01 | 81.19 | ||||||||
| 9 | Zoff239268 | Ⅰ | 6.08 | 68 096.41 | 629 | 2.07 | 11.76 | 86.17 | ||||||||
| 10 | Zoff244943 | Ⅰ | 5.93 | 53 600.22 | 492 | 4.27 | 14.02 | 81.71 | ||||||||
| 11 | Zoff253493 | Ⅰ | 8.78 | 59 842.01 | 551 | 1.63 | 12.89 | 85.48 | ||||||||
| 12 | Zoff258147 | Ⅰ | 6.26 | 78 055.61 | 718 | 3.48 | 11.56 | 84.96 | ||||||||
| 13 | Zoff284143 | Ⅰ | 6.70 | 53 481.19 | 491 | 4.07 | 15.07 | 80.86 | ||||||||
| 14 | Zoff295642 | Ⅰ | 5.41 | 75 059.16 | 690 | 3.19 | 11.01 | 85.80 | ||||||||
| 15 | Zoff193573 | Ⅱ-a | 7.66 | 17 444.56 | 151 | 27.15 | 12.58 | 60.26 | ||||||||
| 16 | Zoff197334 | Ⅱ-a | 9.74 | 32 649.05 | 307 | 7.82 | 13.68 | 78.50 | ||||||||
| 17 | Zoff202059 | Ⅱ-a | 8.77 | 23 689.98 | 208 | 22.12 | 16.83 | 61.06 | ||||||||
| 18 | Zoff204548 | Ⅱ-a | 9.33 | 24 747.76 | 218 | 20.64 | 14.22 | 65.14 | ||||||||
| 19 | Zoff224514 | Ⅱ-a | 9.18 | 22 334.06 | 198 | 16.67 | 13.64 | 69.70 | ||||||||
| 20 | Zoff224526 | Ⅱ-a | 6.72 | 22 023.94 | 199 | 21.11 | 16.58 | 62.31 | ||||||||
| 序号 Number | 序列ID Sequence ID | 类型 Type | 理论等电点 PI | 相对分子质量 MW (kDa) | 氨基酸长度 Amino acid length | α-螺旋 α-helix (%) | β-折叠 β-fold (%) | 其他 Others (%) | ||||||||
| 21 | Zoff224527 | Ⅱ-a | 8.64 | 12 187.19 | 107 | 0.00 | 28.04 | 71.96 | ||||||||
| 22 | Zoff237180 | Ⅱ-a | 9.26 | 23 765.91 | 206 | 21.36 | 13.11 | 65.53 | ||||||||
| 23 | Zoff296350 | Ⅱ-a | 8.76 | 33 220.01 | 303 | 14.52 | 18.48 | 67.00 | ||||||||
| 24 | Zoff600680 | Ⅱ-a | 8.61 | 22 434.71 | 200 | 0.00 | 18.50 | 81.50 | ||||||||
| 25 | Zoff025802 | Ⅱ-b | 8.49 | 34 543.18 | 310 | 11.29 | 13.87 | 74.84 | ||||||||
| 26 | Zoff192125 | Ⅱ-b | 8.78 | 25 559.49 | 242 | 11.16 | 14.88 | 73.97 | ||||||||
| 27 | Zoff199392 | Ⅱ-b | 8.74 | 40 995.52 | 370 | 15.95 | 16.22 | 67.84 | ||||||||
| 28 | Zoff211883 | Ⅱ-b | 8.69 | 30 587.88 | 279 | 3.23 | 14.34 | 82.44 | ||||||||
| 29 | Zoff228302 | Ⅱ-b | 7.79 | 39 532.76 | 353 | 18.98 | 8.78 | 72.24 | ||||||||
| 30 | Zoff229635 | Ⅱ-b | 6.97 | 42 701.98 | 392 | 8.42 | 11.48 | 80.10 | ||||||||
| 31 | Zoff237181 | Ⅱ-b | 8.14 | 39 597.65 | 367 | 16.62 | 11.72 | 71.66 | ||||||||
| 32 | Zoff244550 | Ⅱ-b | 9.31 | 49 121.07 | 456 | 18.20 | 10.96 | 70.83 | ||||||||
| 33 | Zoff255712 | Ⅱ-b | 5.85 | 44 598.95 | 418 | 16.51 | 13.64 | 69.86 | ||||||||
| 34 | Zoff275612 | Ⅱ-b | 8.28 | 23 926.18 | 210 | 20.95 | 9.05 | 70.00 | ||||||||
| 35 | Zoff290487 | Ⅱ-b | 8.99 | 13 316.46 | 122 | 0.82 | 13.11 | 86.07 | ||||||||
| 36 | Zoff290491 | Ⅱ-b | 6.07 | 35 980.81 | 324 | 15.43 | 9.88 | 74.69 | ||||||||
| 37 | Zoff129734 | Ⅱ-c | 9.79 | 17 944.08 | 164 | 6.71 | 18.29 | 75.00 | ||||||||
| 38 | Zoff147627 | Ⅱ-c | 10.04 | 24 896.67 | 223 | 5.38 | 16.59 | 78.03 | ||||||||
| 39 | Zoff165467 | Ⅱ-c | 6.65 | 21 534.40 | 190 | 6.32 | 19.47 | 74.21 | ||||||||
| 40 | Zoff173677 | Ⅱ-c | 9.20 | 33 840.50 | 317 | 6.62 | 14.83 | 78.55 | ||||||||
| 41 | Zoff186957 | Ⅱ-c | 9.60 | 21 486.76 | 187 | 0.00 | 19.79 | 80.21 | ||||||||
| 42 | Zoff226774 | Ⅱ-c | 5.96 | 40 489.58 | 368 | 2.45 | 11.41 | 86.14 | ||||||||
| 43 | Zoff239221 | Ⅱ-c | 9.51 | 21 050.93 | 191 | 2.62 | 19.37 | 78.01 | ||||||||
| 44 | Zoff242316 | Ⅱ-c | 9.51 | 16 605.34 | 149 | 0.00 | 22.82 | 77.18 | ||||||||
| 45 | Zoff253853 | Ⅱ-c | 6.01 | 30 576.87 | 262 | 0.00 | 15.27 | 84.73 | ||||||||
| 46 | Zoff290286 | Ⅱ-c | 9.93 | 19 356.01 | 171 | 4.09 | 21.64 | 74.27 | ||||||||
| 47 | Zoff614319 | Ⅱ-c | 10.14 | 12 362.28 | 109 | 0.00 | 33.94 | 66.06 | ||||||||
| 48 | Zoff150646 | Ⅱ-d | 9.87 | 29 012.97 | 269 | 7.06 | 15.24 | 77.70 | ||||||||
| 49 | Zoff170651 | Ⅱ-d | 10.12 | 34 732.26 | 321 | 22.12 | 10.59 | 67.29 | ||||||||
| 50 | Zoff185798 | Ⅱ-d | 9.98 | 27 468.14 | 252 | 23.41 | 15.08 | 61.51 | ||||||||
| 51 | Zoff186024 | Ⅱ-d | 9.55 | 41 308.75 | 372 | 14.78 | 11.56 | 73.66 | ||||||||
| 52 | Zoff188265 | Ⅱ-d | 9.75 | 39 505.83 | 352 | 12.78 | 12.22 | 75.00 | ||||||||
| 53 | Zoff192006 | Ⅱ-d | 10.18 | 33 047.28 | 308 | 10.71 | 14.29 | 75.00 | ||||||||
| 54 | Zoff199154 | Ⅱ-d | 9.85 | 38 393.36 | 346 | 12.43 | 12.14 | 75.43 | ||||||||
| 55 | Zoff204858 | Ⅱ-d | 9.77 | 35 680.37 | 324 | 16.98 | 12.65 | 70.37 | ||||||||
| 56 | Zoff208722 | Ⅱ-d | 10.15 | 12 913.60 | 116 | 0.00 | 33.62 | 66.38 | ||||||||
| 57 | Zoff209584 | Ⅱ-d | 9.95 | 35 415.23 | 328 | 15.85 | 11.89 | 72.26 | ||||||||
| 58 | Zoff238579 | Ⅱ-d | 9.63 | 38 150.32 | 344 | 12.50 | 11.92 | 75.58 | ||||||||
| 59 | Zoff238581 | Ⅱ-d | 9.71 | 38 594.60 | 345 | 12.75 | 12.17 | 75.07 | ||||||||
| 60 | Zoff305561 | Ⅱ-d | 9.90 | 28 447.66 | 271 | 19.93 | 7.01 | 73.06 | ||||||||
| 61 | Zoff407487 | Ⅱ-d | 10.13 | 14 806.36 | 136 | 0.00 | 15.44 | 84.56 | ||||||||
| 62 | Zoff066597 | Ⅱ-e | 10.86 | 12 246.39 | 108 | 0.93 | 30.56 | 68.52 | ||||||||
| 63 | Zoff178498 | Ⅱ-e | 9.69 | 26 402.19 | 236 | 2.54 | 17.37 | 80.08 | ||||||||
| 64 | Zoff186238 | Ⅱ-e | 8.34 | 23 262.53 | 212 | 3.77 | 18.40 | 77.83 | ||||||||
| 65 | Zoff223289 | Ⅱ-e | 9.23 | 23 531.70 | 214 | 6.54 | 22.43 | 71.03 | ||||||||
| 66 | Zoff288806 | Ⅱ-e | 7.00 | 17 884.78 | 160 | 3.75 | 15.00 | 81.25 | ||||||||
| 67 | Zoff288813 | Ⅱ-e | 5.39 | 41 512.52 | 384 | 0.78 | 12.24 | 86.98 | ||||||||
| 68 | Zoff311150 | Ⅱ-e | 5.77 | 35 150.65 | 326 | 7.98 | 11.66 | 80.37 | ||||||||
| 69 | Zoff536230 | Ⅱ-e | 9.41 | 16 935.56 | 148 | 6.08 | 14.86 | 79.05 | ||||||||
| 70 | Zoff109660 | Ⅲ | 7.71 | 29 460.72 | 261 | 21.46 | 9.96 | 68.58 | ||||||||
| 71 | Zoff160883 | Ⅲ | 9.23 | 12 035.04 | 102 | 21.57 | 34.31 | 44.12 | ||||||||
| 72 | Zoff184272 | Ⅲ | 9.35 | 24 075.94 | 215 | 26.51 | 16.28 | 57.21 | ||||||||
| 73 | Zoff188162 | Ⅲ | 9.89 | 27 125.18 | 243 | 21.81 | 10.29 | 67.90 | ||||||||
| 74 | Zoff196199 | Ⅲ | 5.96 | 39 530.50 | 355 | 16.90 | 9.86 | 73.24 | ||||||||
| 75 | Zoff217771 | Ⅲ | 6.23 | 32 122.87 | 290 | 13.45 | 13.79 | 72.76 | ||||||||
| 76 | Zoff254593 | Ⅲ | 8.79 | 20 719.11 | 183 | 3.28 | 19.13 | 77.60 | ||||||||
| 77 | Zoff301103 | Ⅲ | 5.68 | 24 958.96 | 224 | 4.91 | 15.18 | 79.91 | ||||||||
| 78 | Zoff554263 | Ⅲ | 9.79 | 11 591.13 | 104 | 30.77 | 8.65 | 60.58 | ||||||||
通过Sopma网站预测蛋白质二级结构,ZoWRKY成员中α-螺旋、β-折叠和无规则卷曲等结构在全序列中的占比差异较大,但构成元件中主要为无规则卷曲,平均占比大于70%。亚族Ⅰ的α-螺旋占比最低,平均为2.63%;亚族Ⅲ的α-螺旋占比最高,平均为17.85%。
2.6 ZoWRKY家族对逆境的响应模式
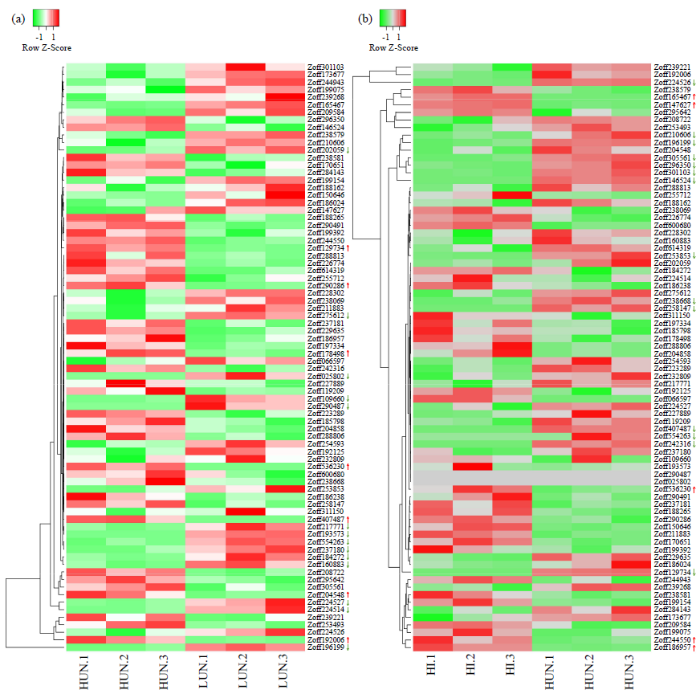
基于不同WFPS(10%、25%、30%和40%)及青枯菌侵染处理的生姜根茎转录组数据,分析ZoWRKY基因对逆境胁迫的响应。Zoff129734等7个ZoWRKY基因在高湿度土壤中上调表达,主要分布在亚族Ⅱ-c和Ⅱ-d,其中Zoff204548、Zoff129734、Zoff290286、Zoff407487和Zoff178498分别上调了2.89、12.52、4.20、3.66和5.26倍。而Zoff193573等14个ZoWRKY基因下调表达,主要分布在亚族Ⅱ-a、Ⅱ-b和Ⅲ中,其中Zoff193573、Zoff224514、Zoff224527、Zoff109660和Zoff554263分别下调至0.02、0.06、0.06、0.03和0.04(图4a)。
图4
图4
ZoWRKY基因对逆境的响应分析
(a) ZoWRKY基因对土壤湿度的响应。(b) ZoWRKY基因对青枯菌侵染的响应。LUN:低土壤湿度(WFPS:10%),HUN:高土壤湿度(WFPS:40%),HI:高土壤湿度(WFPS:40%)且感染青枯菌;根据每个ZoWRKY基因的表达水平分组计算log2FC,通过edgeR对2组样本间的差异基因进行识别,阈值为log2FC≥1和FDR < 0.05;色阶标尺从左-1.0到右1.0逐渐增大;右侧红色箭头表示上调表达,绿色箭头表示下调表达
Fig.4
Expression analysis of ZoWRKY gene under stress conditions
(a) Gene ZoWRKY responses to soil moistures. (b) Gene ZoWRKY responses to infection of R. solanacearum. LUN: low soil moisture (WFPS: 10%); HUN: high soil moisture (WFPS: 40%); HI: high soil moisture (WFPS: 40%) and infection of R. solanacearum; log2FC value was calculated pairwise based on the expression level for each ZoWRKY gene, DEGs were identified between each two groups by edgeR with thresholds of log2FC ≥1 and FDR < 0.05; color scale increases from left to right with values -1.0 to 1.0; red arrow indicates up-regulated expression, green arrow indicates down-regulated expression
青枯菌侵染分析发现,Zoff186957等19个ZoWRKY基因显著响应青枯菌侵染,其中在侵染过程中显著上调表达的5个主要分布亚族Ⅱ-b和Ⅱ-c中,Zoff147627、Zoff165467、Zoff186957、Zoff244550和Zoff536230分别上调了7.03、3.44、37.62、2.87和7.38倍。而Zoff129734等14个ZoWRKY基因下调表达,主要分布在亚族Ⅰ、Ⅱ-a和Ⅲ中,其中Zoff129734、Zoff146524、Zoff224526、Zoff258147和Zoff301103分别下调至0.02、0.07、0.17、0.03和0.09(图4b)。
3 讨论
WRKY家族成员在植物体内行使多种功能,当植物感知生物或非生物逆境胁迫后,可能启动一系列ZoWRKY表达,选择多种内源激素介导的信号通路,进而调控抗性基因转录,在抗性和防御反应中发挥作用。生姜中ZoWRKY成员分别有21个响应土壤湿度和19个响应青枯菌侵染,在Ⅱ-c有共同上调表达的成员,在Ⅱ-a和Ⅲ有共同下调表达的成员。高土壤湿度下呈上调表达的7个ZoWRKY主要聚类在Ⅱ-c和Ⅱ-d,结合与拟南芥同源性较高的AtWRKY8和AtWRKY17功能,推测其主要调控逆境信号通路和胁迫耐受[29]。下调表达的14个ZoWRKY主要分布在Ⅱ-a、Ⅱ-b和Ⅲ,结合同源性较高的AtWRKY40、AtWRKY6及AtWRKY54等功能,推测其主要调节防御、衰老和气孔运动等过程[30];青枯菌侵染引起上调表达的5个ZoWRKY基因主要聚类在Ⅱ-b和Ⅱ-c,结合同源性较高的AtWRKY6和AtWRKY51的功能,推测其主要调控病原体防御和防御反应信号通路[31]。下调表达的14个ZoWRKY主要聚类在Ⅰ、Ⅱ-a和Ⅲ,结合同源性较高的AtWRKY26、AtWRKY18和AtWRKY70等功能,推测其主要参与细胞分化、防御和信号通路选择[32-33]。
4 结论
WRKY作为植物特有的转录因子家族,在生长发育、信号传导和逆境胁迫应答中发挥着重要作用,但关于ZoWRKY转录因子鉴定及逆境响应分析还未见报道。本研究借助WRKY保守域种子序列检索生姜Unigenes编码蛋白质数据库,共筛选鉴定了78个ZoWRKY成员,分为3个亚族,其中有21个和19个ZoWRKY基因分别响应不同WFPS和青枯菌侵染,参与调节逆境胁迫下生姜生长发育和对病原菌的防御反应等过程。结果有助于进一步探究ZoWRKY家族的生物学功能,解析生姜响应环境变化如何调控ZoWRKY基因转录表达,对生姜的抗性育种、提高产量及品质具有重要的意义。
参考文献
MYB transcription factors that colour our fruit
DOI:10.1016/j.tplants.2007.11.012 URL [本文引用: 1]
MYB transcription factors as regulators of phenylpropanoid metabolism in plants
DOI:10.1016/j.molp.2015.03.012 URL [本文引用: 1]
Involvement of CBF transcription factors in winter hardiness in birch
DOI:10.1104/pp.108.117812 URL [本文引用: 1]
Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis
DOI:10.1104/pp.16.00533 URL [本文引用: 1]
NAC transcription factors:structurally distinct,functionally diverse
DOI:10.1016/j.tplants.2004.12.010 URL [本文引用: 1]
NAC-type transcription factors regulate accumulation of starch and protein in maize seeds
植物WRKY转录因子家族研究进展
DOI:10.13560/j.cnki.biotech.bull.1985.2019-0626
[本文引用: 1]

WRKY转录因子是植物中最大的转录调控因子家族之一,是调控植物许多生物过程信号网络的组成部分。WKRY转录因子具有多种生物学功能,在植物的生长发育和衰老、非生物和生物胁迫等过程中发挥着重要的作用。在DNA水平上,WRKY转录因子可与靶基因启动子中的W-box TTGAC(C/T)结合,通过自调节或交叉调节激活或抑制下游基因的表达调控其反应。在蛋白水平上,WRKY转录因子可以与多种蛋白相互作用,包括MAP激酶、组蛋白去乙酰化酶、抗性R蛋白、多种转录因子等,调节植物的生长发育或各种应激反应。对WRKY转录因子的结构特征、生物学功能、调控机制和网络等方面进行了综述,有助于更加全面了解其在植物中的作用。
The WRKY family of transcription factors in rice and Arabidopsis and their origins
DOI:10.1093/dnares/12.1.9 URL [本文引用: 1]
WRKY transcription factors
DOI:10.1016/j.tplants.2010.02.006
PMID:20304701
[本文引用: 1]

WRKY transcription factors are one of the largest families of transcriptional regulators in plants and form integral parts of signalling webs that modulate many plant processes. Here, we review recent significant progress in WRKY transcription factor research. New findings illustrate that WRKY proteins often act as repressors as well as activators, and that members of the family play roles in both the repression and de-repression of important plant processes. Furthermore, it is becoming clear that a single WRKY transcription factor might be involved in regulating several seemingly disparate processes. Mechanisms of signalling and transcriptional regulation are being dissected, uncovering WRKY protein functions via interactions with a diverse array of protein partners, including MAP kinases, MAP kinase kinases, 14-3-3 proteins, calmodulin, histone deacetylases, resistance proteins and other WRKY transcription factors. WRKY genes exhibit extensive autoregulation and cross-regulation that facilitates transcriptional reprogramming in a dynamic web with built-in redundancy.2010 Elsevier Ltd. All rights reserved.
The WRKY superfamily of plant transcription factors
The WRKY proteins are a superfamily of transcription factors with up to 100 representatives in Arabidopsis. Family members appear to be involved in the regulation of various physio-logical programs that are unique to plants, including pathogen defense, senescence and trichome development. In spite of the strong conservation of their DNA-binding domain, the overall structures of WRKY proteins are highly divergent and can be categorized into distinct groups, which might reflect their different functions.
WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis
WRKY transcription factors in plant responses to stresses
DOI:10.1111/jipb.12513 URL [本文引用: 1]
CaWRKY40,a WRKY protein of pepper,plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection
DOI:10.1111/pce.12011 URL [本文引用: 1]
The transcription factor VvWRKY 33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmopara viticola
DOI:10.1111/ppl.12251 URL [本文引用: 1]
Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley
WRKY transcription factors:Jack of many trades in plants
DOI:10.4161/psb.27700 URL [本文引用: 2]
CaWRKY6 transcriptionally activates CaWRKY40,regulates Ralstonia solanacearum resistance,and confers high-temperature and high-humidity tolerance in pepper
DOI:10.1093/jxb/erv125 URL [本文引用: 1]
WRKY transcription factors:from DNA binding towards biological function
DOI:10.1016/j.pbi.2004.07.012 URL [本文引用: 2]
The WRKY gene family in rice (Oryza sativa)
DOI:10.1111/j.1744-7909.2007.00504.x URL [本文引用: 1]
Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum
DOI:10.1007/s00438-012-0696-6 URL [本文引用: 2]
Genome-wide analysis of the WRKY gene family in cotton
DOI:10.1007/s00438-014-0872-y URL [本文引用: 1]
Gingerols and shogaols:important nutraceutical principles from ginger
DOI:10.1016/j.phytochem.2015.07.012 URL [本文引用: 1]
A novel WRKY transcription factor,SUSIBA2,participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter
DOI:10.1105/tpc.014597 URL [本文引用: 1]
Networks of WRKY transcription factors in defense signaling
DOI:10.1016/j.pbi.2007.04.020 URL [本文引用: 1]
Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis
DOI:10.1016/j.jplph.2018.04.007 URL [本文引用: 1]
WRKY70 modulates the selection of signaling pathways in plant defense
DOI:10.1111/j.1365-313X.2006.02712.x URL [本文引用: 1]
Overexpression of AtWRKY28 and AtWRKY 75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum
DOI:10.1007/s00299-013-1469-3 URL [本文引用: 1]
Overexpression of AtWRKY 30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana
DOI:10.1007/s11103-013-0090-8
PMID:23794142
[本文引用: 1]

AtWRKY30 belongs to a higher plant transcription factor superfamily, which responds to pathogen attack. In previous studies, the AtWRKY30 gene was found to be highly and rapidly induced in Arabidopsis thaliana leaves after oxidative stress treatment. In this study, electrophoretic mobility shift assays showed that AtWRKY30 binds with high specificity and affinity to the WRKY consensus sequence (W-box), and also to its own promoter. Analysis of the AtWRKY30 expression pattern by qPCR and using transgenic Arabidopsis lines carrying AtWRKY30 promoter-β-glucuronidase fusions showed transcriptional activity in leaves subjected to biotic or abiotic stress. Transgenic Arabidopsis plants constitutively overexpressing AtWRKY30 (35S::W30 lines) were more tolerant than wild-type plants to oxidative and salinity stresses during seed germination. The results presented here show that AtWRKY30 is responsive to several stress conditions either from abiotic or biotic origin, suggesting that AtWRKY30 could have a role in the activation of defence responses at early stages of Arabidopsis growth by binding to W-boxes found in promoters of many stress/developmentally regulated genes.
Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40,and WRKY 60 transcription factors
DOI:10.1105/tpc.105.037523 URL [本文引用: 1]